Giosue Scognamiglio
HACT-Net: A Hierarchical Cell-to-Tissue Graph Neural Network for Histopathological Image Classification
Jul 01, 2020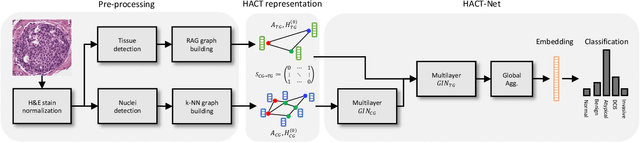
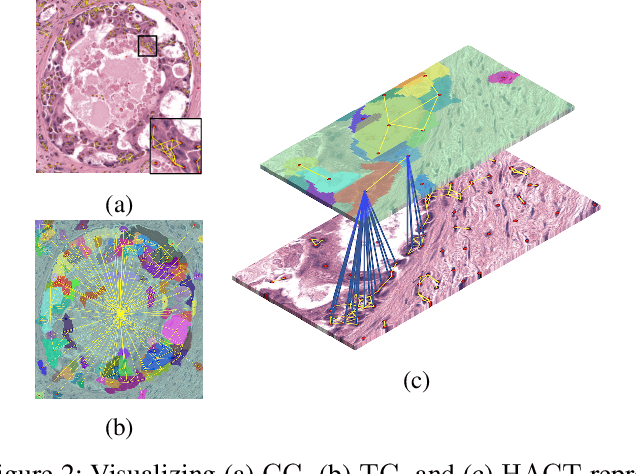
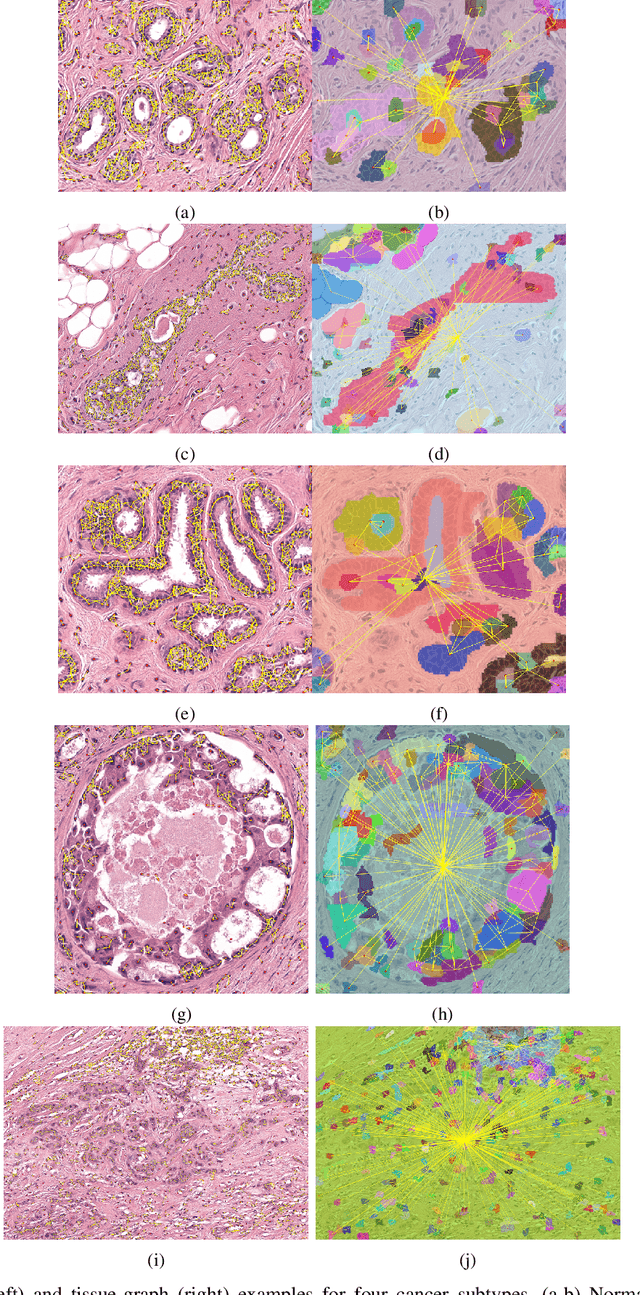

Abstract:Cancer diagnosis, prognosis, and therapeutic response prediction are heavily influenced by the relationship between the histopathological structures and the function of the tissue. Recent approaches acknowledging the structure-function relationship, have linked the structural and spatial patterns of cell organization in tissue via cell-graphs to tumor grades. Though cell organization is imperative, it is insufficient to entirely represent the histopathological structure. We propose a novel hierarchical cell-to-tissue-graph (HACT) representation to improve the structural depiction of the tissue. It consists of a low-level cell-graph, capturing cell morphology and interactions, a high-level tissue-graph, capturing morphology and spatial distribution of tissue parts, and cells-to-tissue hierarchies, encoding the relative spatial distribution of the cells with respect to the tissue distribution. Further, a hierarchical graph neural network (HACT-Net) is proposed to efficiently map the HACT representations to histopathological breast cancer subtypes. We assess the methodology on a large set of annotated tissue regions of interest from H\&E stained breast carcinoma whole-slides. Upon evaluation, the proposed method outperformed recent convolutional neural network and graph neural network approaches for breast cancer multi-class subtyping. The proposed entity-based topological analysis is more inline with the pathological diagnostic procedure of the tissue. It provides more command over the tissue modelling, therefore encourages the further inclusion of pathological priors into task-specific tissue representation.
Towards Explainable Graph Representations in Digital Pathology
Jul 01, 2020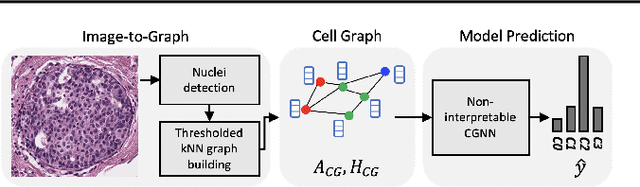
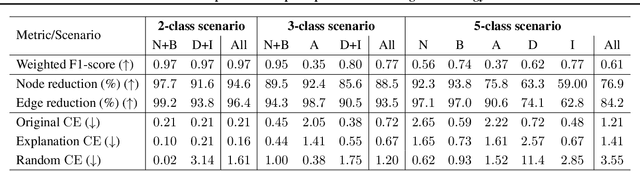
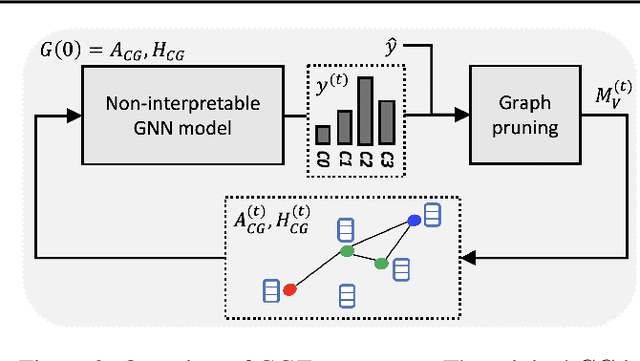
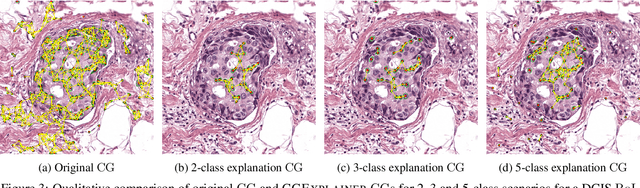
Abstract:Explainability of machine learning (ML) techniques in digital pathology (DP) is of great significance to facilitate their wide adoption in clinics. Recently, graph techniques encoding relevant biological entities have been employed to represent and assess DP images. Such paradigm shift from pixel-wise to entity-wise analysis provides more control over concept representation. In this paper, we introduce a post-hoc explainer to derive compact per-instance explanations emphasizing diagnostically important entities in the graph. Although we focus our analyses to cells and cellular interactions in breast cancer subtyping, the proposed explainer is generic enough to be extended to other topological representations in DP. Qualitative and quantitative analyses demonstrate the efficacy of the explainer in generating comprehensive and compact explanations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge