Giuseppe De Pietro
Quantum Transfer Learning for Acceptability Judgements
Jan 15, 2024Abstract:Hybrid quantum-classical classifiers promise to positively impact critical aspects of natural language processing tasks, particularly classification-related ones. Among the possibilities currently investigated, quantum transfer learning, i.e., using a quantum circuit for fine-tuning pre-trained classical models for a specific task, is attracting significant attention as a potential platform for proving quantum advantage. This work shows potential advantages, both in terms of performance and expressiveness, of quantum transfer learning algorithms trained on embedding vectors extracted from a large language model to perform classification on a classical Linguistics task: acceptability judgments. Acceptability judgment is the ability to determine whether a sentence is considered natural and well-formed by a native speaker. The approach has been tested on sentences extracted from ItaCoLa, a corpus that collects Italian sentences labeled with their acceptability judgment. The evaluation phase shows results for the quantum transfer learning pipeline comparable to state-of-the-art classical transfer learning algorithms, proving current quantum computers' capabilities to tackle NLP tasks for ready-to-use applications. Furthermore, a qualitative linguistic analysis, aided by explainable AI methods, reveals the capabilities of quantum transfer learning algorithms to correctly classify complex and more structured sentences, compared to their classical counterpart. This finding sets the ground for a quantifiable quantum advantage in NLP in the near future.
BRACS: A Dataset for BReAst Carcinoma Subtyping in H&E Histology Images
Nov 08, 2021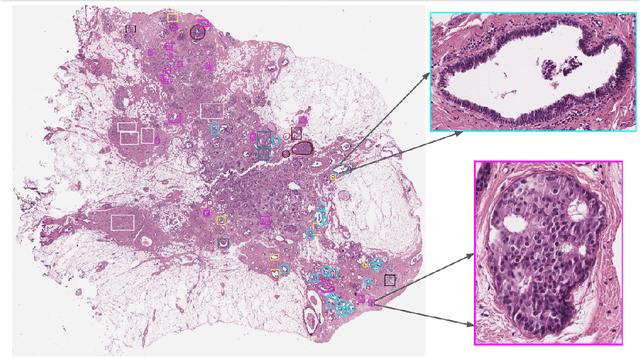

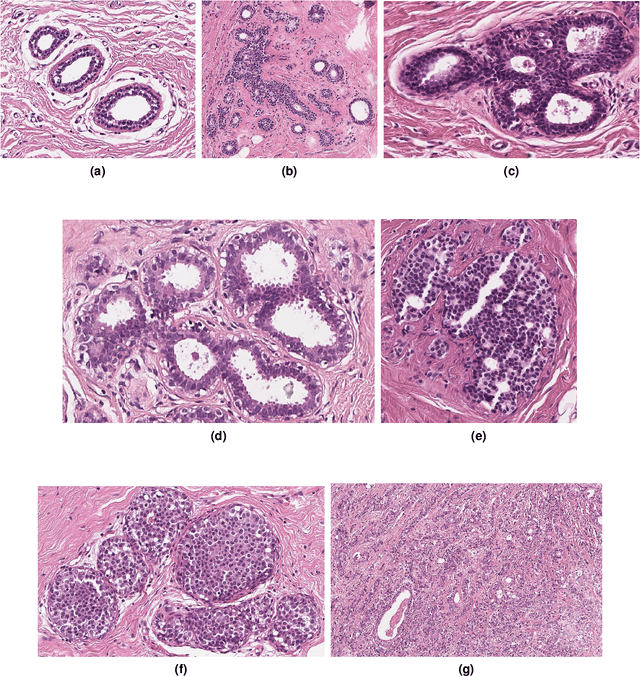
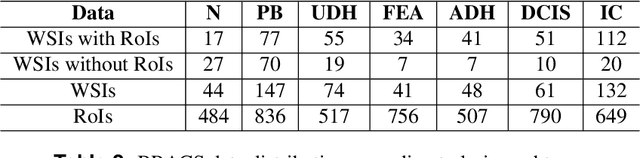
Abstract:Breast cancer is the most commonly diagnosed cancer and registers the highest number of deaths for women with cancer. Recent advancements in diagnostic activities combined with large-scale screening policies have significantly lowered the mortality rates for breast cancer patients. However, the manual inspection of tissue slides by the pathologists is cumbersome, time-consuming, and is subject to significant inter- and intra-observer variability. Recently, the advent of whole-slide scanning systems have empowered the rapid digitization of pathology slides, and enabled to develop digital workflows. These advances further enable to leverage Artificial Intelligence (AI) to assist, automate, and augment pathological diagnosis. But the AI techniques, especially Deep Learning (DL), require a large amount of high-quality annotated data to learn from. Constructing such task-specific datasets poses several challenges, such as, data-acquisition level constrains, time-consuming and expensive annotations, and anonymization of private information. In this paper, we introduce the BReAst Carcinoma Subtyping (BRACS) dataset, a large cohort of annotated Hematoxylin & Eosin (H&E)-stained images to facilitate the characterization of breast lesions. BRACS contains 547 Whole-Slide Images (WSIs), and 4539 Regions of Interest (ROIs) extracted from the WSIs. Each WSI, and respective ROIs, are annotated by the consensus of three board-certified pathologists into different lesion categories. Specifically, BRACS includes three lesion types, i.e., benign, malignant and atypical, which are further subtyped into seven categories. It is, to the best of our knowledge, the largest annotated dataset for breast cancer subtyping both at WSI- and ROI-level. Further, by including the understudied atypical lesions, BRACS offers an unique opportunity for leveraging AI to better understand their characteristics.
Hierarchical Graph Representations in Digital Pathology
Mar 17, 2021



Abstract:Cancer diagnosis, prognosis, and therapy response predictions from tissue specimens highly depend on the phenotype and topological distribution of constituting histological entities. Thus, adequate tissue representations for encoding histological entities is imperative for computer aided cancer patient care. To this end, several approaches have leveraged cell-graphs that encode cell morphology and organization to denote the tissue information. These allow for utilizing machine learning to map tissue representations to tissue functionality to help quantify their relationship. Though cellular information is crucial, it is incomplete alone to comprehensively characterize complex tissue structure. We herein treat the tissue as a hierarchical composition of multiple types of histological entities from fine to coarse level, capturing multivariate tissue information at multiple levels. We propose a novel multi-level hierarchical entity-graph representation of tissue specimens to model hierarchical compositions that encode histological entities as well as their intra- and inter-entity level interactions. Subsequently, a graph neural network is proposed to operate on the hierarchical entity-graph representation to map the tissue structure to tissue functionality. Specifically, for input histology images we utilize well-defined cells and tissue regions to build HierArchical Cell-to-Tissue (HACT) graph representations, and devise HACT-Net, a graph neural network, to classify such HACT representations. As part of this work, we introduce the BReAst Carcinoma Subtyping (BRACS) dataset, a large cohort of H&E stained breast tumor images, to evaluate our proposed methodology against pathologists and state-of-the-art approaches. Through comparative assessment and ablation studies, our method is demonstrated to yield superior classification results compared to alternative methods as well as pathologists.
Gigapixel Histopathological Image Analysis using Attention-based Neural Networks
Jan 30, 2021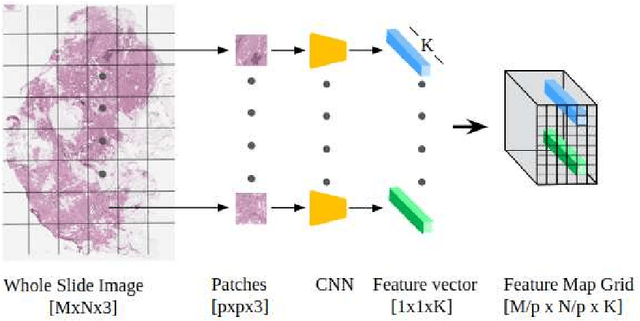
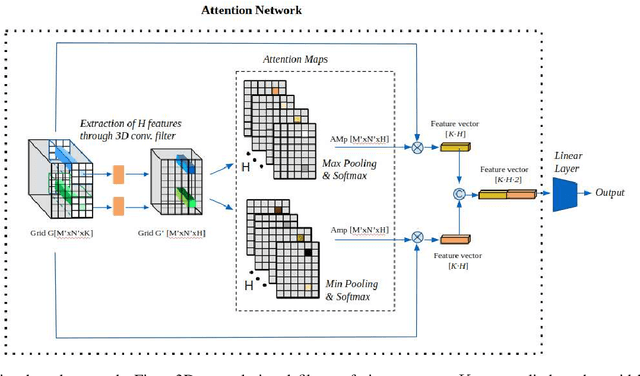
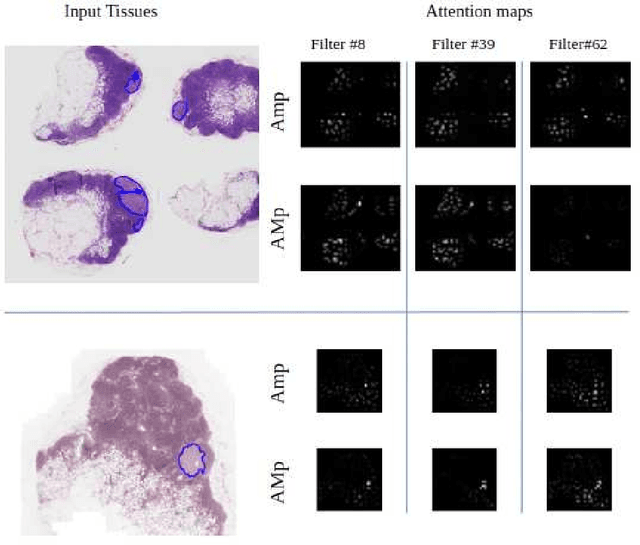
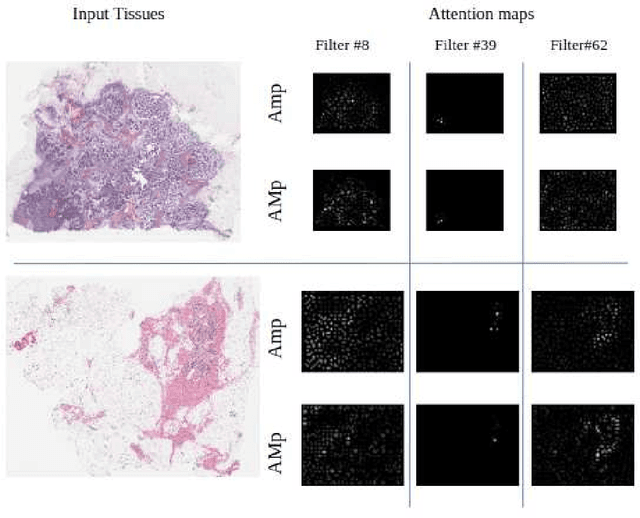
Abstract:Although CNNs are widely considered as the state-of-the-art models in various applications of image analysis, one of the main challenges still open is the training of a CNN on high resolution images. Different strategies have been proposed involving either a rescaling of the image or an individual processing of parts of the image. Such strategies cannot be applied to images, such as gigapixel histopathological images, for which a high reduction in resolution inherently effects a loss of discriminative information, and in respect of which the analysis of single parts of the image suffers from a lack of global information or implies a high workload in terms of annotating the training images in such a way as to select significant parts. We propose a method for the analysis of gigapixel histopathological images solely by using weak image-level labels. In particular, two analysis tasks are taken into account: a binary classification and a prediction of the tumor proliferation score. Our method is based on a CNN structure consisting of a compressing path and a learning path. In the compressing path, the gigapixel image is packed into a grid-based feature map by using a residual network devoted to the feature extraction of each patch into which the image has been divided. In the learning path, attention modules are applied to the grid-based feature map, taking into account spatial correlations of neighboring patch features to find regions of interest, which are then used for the final whole slide analysis. Our method integrates both global and local information, is flexible with regard to the size of the input images and only requires weak image-level labels. Comparisons with different methods of the state-of-the-art on two well known datasets, Camelyon16 and TUPAC16, have been made to confirm the validity of the proposed model.
HACT-Net: A Hierarchical Cell-to-Tissue Graph Neural Network for Histopathological Image Classification
Jul 01, 2020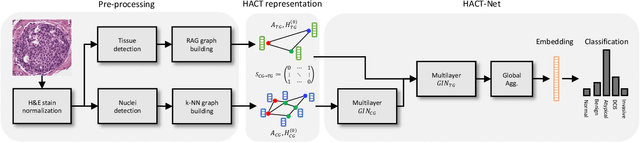
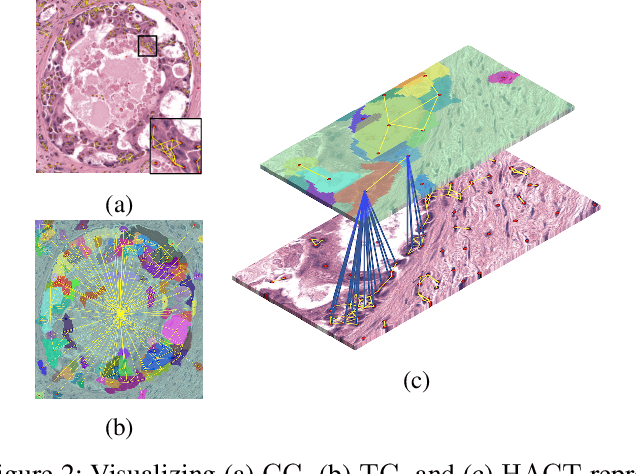
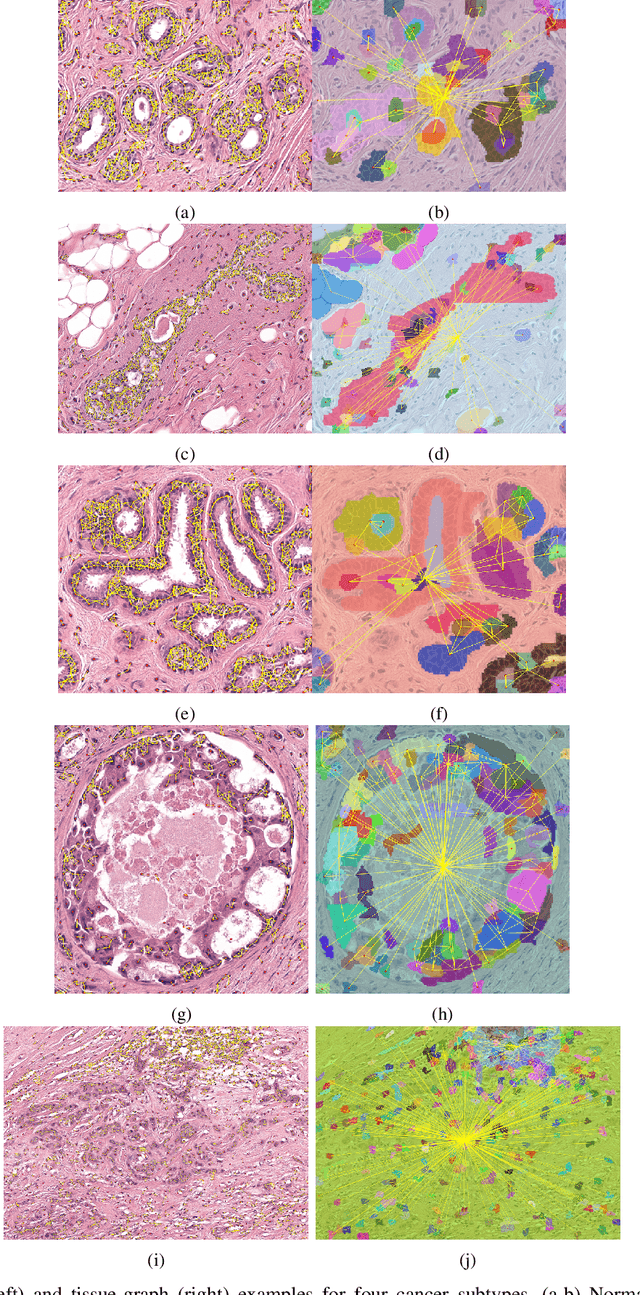

Abstract:Cancer diagnosis, prognosis, and therapeutic response prediction are heavily influenced by the relationship between the histopathological structures and the function of the tissue. Recent approaches acknowledging the structure-function relationship, have linked the structural and spatial patterns of cell organization in tissue via cell-graphs to tumor grades. Though cell organization is imperative, it is insufficient to entirely represent the histopathological structure. We propose a novel hierarchical cell-to-tissue-graph (HACT) representation to improve the structural depiction of the tissue. It consists of a low-level cell-graph, capturing cell morphology and interactions, a high-level tissue-graph, capturing morphology and spatial distribution of tissue parts, and cells-to-tissue hierarchies, encoding the relative spatial distribution of the cells with respect to the tissue distribution. Further, a hierarchical graph neural network (HACT-Net) is proposed to efficiently map the HACT representations to histopathological breast cancer subtypes. We assess the methodology on a large set of annotated tissue regions of interest from H\&E stained breast carcinoma whole-slides. Upon evaluation, the proposed method outperformed recent convolutional neural network and graph neural network approaches for breast cancer multi-class subtyping. The proposed entity-based topological analysis is more inline with the pathological diagnostic procedure of the tissue. It provides more command over the tissue modelling, therefore encourages the further inclusion of pathological priors into task-specific tissue representation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge