Kristin P. Bennett
Consistent Causal Inference of Group Effects in Non-Targeted Trials with Finitely Many Effect Levels
Apr 22, 2025Abstract:A treatment may be appropriate for some group (the ``sick" group) on whom it has a positive effect, but it can also have a detrimental effect on subjects from another group (the ``healthy" group). In a non-targeted trial both sick and healthy subjects may be treated, producing heterogeneous effects within the treated group. Inferring the correct treatment effect on the sick population is then difficult, because the effects on the different groups get tangled. We propose an efficient nonparametric approach to estimating the group effects, called {\bf PCM} (pre-cluster and merge). We prove its asymptotic consistency in a general setting and show, on synthetic data, more than a 10x improvement in accuracy over existing state-of-the-art. Our approach applies more generally to consistent estimation of functions with a finite range.
CTBench: A Comprehensive Benchmark for Evaluating Language Model Capabilities in Clinical Trial Design
Jun 25, 2024Abstract:CTBench is introduced as a benchmark to assess language models (LMs) in aiding clinical study design. Given study-specific metadata, CTBench evaluates AI models' ability to determine the baseline features of a clinical trial (CT), which include demographic and relevant features collected at the trial's start from all participants. These baseline features, typically presented in CT publications (often as Table 1), are crucial for characterizing study cohorts and validating results. Baseline features, including confounders and covariates, are also necessary for accurate treatment effect estimation in studies involving observational data. CTBench consists of two datasets: "CT-Repo," containing baseline features from 1,690 clinical trials sourced from clinicaltrials.gov, and "CT-Pub," a subset of 100 trials with more comprehensive baseline features gathered from relevant publications. Two LM-based evaluation methods are developed to compare the actual baseline feature lists against LM-generated responses. "ListMatch-LM" and "ListMatch-BERT" use GPT-4o and BERT scores (at various thresholds), respectively, for evaluation. To establish baseline results, advanced prompt engineering techniques using LLaMa3-70B-Instruct and GPT-4o in zero-shot and three-shot learning settings are applied to generate potential baseline features. The performance of GPT-4o as an evaluator is validated through human-in-the-loop evaluations on the CT-Pub dataset, where clinical experts confirm matches between actual and LM-generated features. The results highlight a promising direction with significant potential for improvement, positioning CTBench as a useful tool for advancing research on AI in CT design and potentially enhancing the efficacy and robustness of CTs.
Should we tweet this? Generative response modeling for predicting reception of public health messaging on Twitter
Apr 09, 2022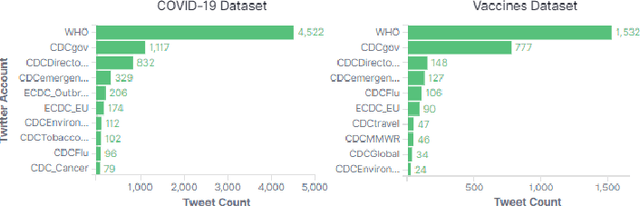
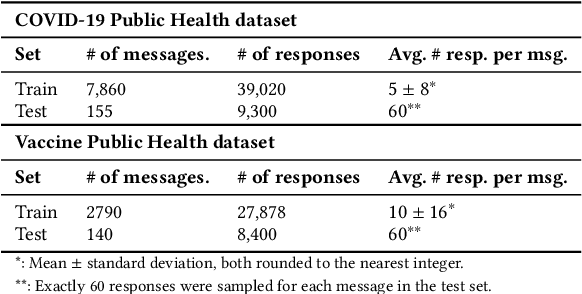
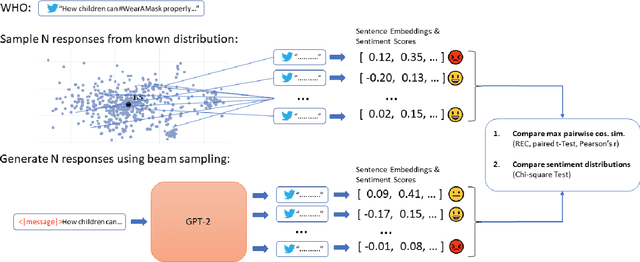
Abstract:The way people respond to messaging from public health organizations on social media can provide insight into public perceptions on critical health issues, especially during a global crisis such as COVID-19. It could be valuable for high-impact organizations such as the US Centers for Disease Control and Prevention (CDC) or the World Health Organization (WHO) to understand how these perceptions impact reception of messaging on health policy recommendations. We collect two datasets of public health messages and their responses from Twitter relating to COVID-19 and Vaccines, and introduce a predictive method which can be used to explore the potential reception of such messages. Specifically, we harness a generative model (GPT-2) to directly predict probable future responses and demonstrate how it can be used to optimize expected reception of important health guidance. Finally, we introduce a novel evaluation scheme with extensive statistical testing which allows us to conclude that our models capture the semantics and sentiment found in actual public health responses.
Downstream Fairness Caveats with Synthetic Healthcare Data
Mar 09, 2022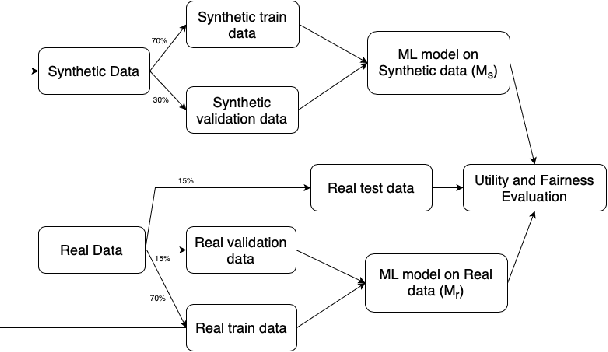

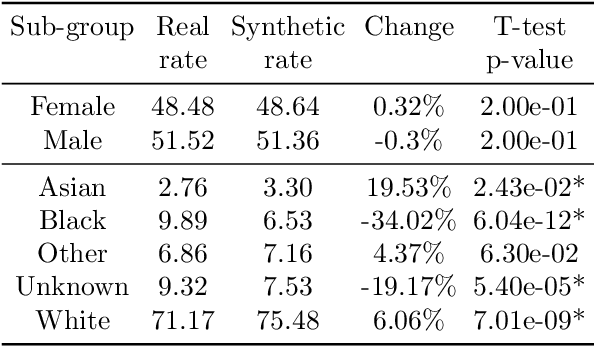
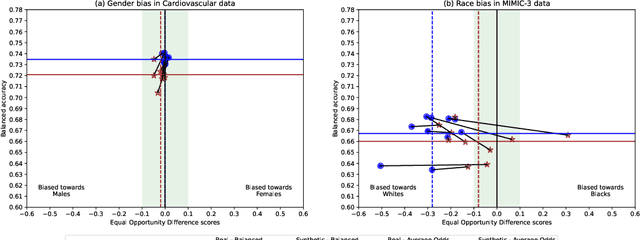
Abstract:This paper evaluates synthetically generated healthcare data for biases and investigates the effect of fairness mitigation techniques on utility-fairness. Privacy laws limit access to health data such as Electronic Medical Records (EMRs) to preserve patient privacy. Albeit essential, these laws hinder research reproducibility. Synthetic data is a viable solution that can enable access to data similar to real healthcare data without privacy risks. Healthcare datasets may have biases in which certain protected groups might experience worse outcomes than others. With the real data having biases, the fairness of synthetically generated health data comes into question. In this paper, we evaluate the fairness of models generated on two healthcare datasets for gender and race biases. We generate synthetic versions of the dataset using a Generative Adversarial Network called HealthGAN, and compare the real and synthetic model's balanced accuracy and fairness scores. We find that synthetic data has different fairness properties compared to real data and fairness mitigation techniques perform differently, highlighting that synthetic data is not bias free.
Synthetic Event Time Series Health Data Generation
Nov 27, 2019



Abstract:Synthetic medical data which preserves privacy while maintaining utility can be used as an alternative to real medical data, which has privacy costs and resource constraints associated with it. At present, most models focus on generating cross-sectional health data which is not necessarily representative of real data. In reality, medical data is longitudinal in nature, with a single patient having multiple health events, non-uniformly distributed throughout their lifetime. These events are influenced by patient covariates such as comorbidities, age group, gender etc. as well as external temporal effects (e.g. flu season). While there exist seminal methods to model time series data, it becomes increasingly challenging to extend these methods to medical event time series data. Due to the complexity of the real data, in which each patient visit is an event, we transform the data by using summary statistics to characterize the events for a fixed set of time intervals, to facilitate analysis and interpretability. We then train a generative adversarial network to generate synthetic data. We demonstrate this approach by generating human sleep patterns, from a publicly available dataset. We empirically evaluate the generated data and show close univariate resemblance between synthetic and real data. However, we also demonstrate how stratification by covariates is required to gain a deeper understanding of synthetic data quality.
Making Study Populations Visible through Knowledge Graphs
Jul 09, 2019



Abstract:Treatment recommendations within Clinical Practice Guidelines (CPGs) are largely based on findings from clinical trials and case studies, referred to here as research studies, that are often based on highly selective clinical populations, referred to here as study cohorts. When medical practitioners apply CPG recommendations, they need to understand how well their patient population matches the characteristics of those in the study cohort, and thus are confronted with the challenges of locating the study cohort information and making an analytic comparison. To address these challenges, we develop an ontology-enabled prototype system, which exposes the population descriptions in research studies in a declarative manner, with the ultimate goal of allowing medical practitioners to better understand the applicability and generalizability of treatment recommendations. We build a Study Cohort Ontology (SCO) to encode the vocabulary of study population descriptions, that are often reported in the first table in the published work, thus they are often referred to as Table 1. We leverage the well-used Semanticscience Integrated Ontology (SIO) for defining property associations between classes. Further, we model the key components of Table 1s, i.e., collections of study subjects, subject characteristics, and statistical measures in RDF knowledge graphs. We design scenarios for medical practitioners to perform population analysis, and generate cohort similarity visualizations to determine the applicability of a study population to the clinical population of interest. Our semantic approach to make study populations visible, by standardized representations of Table 1s, allows users to quickly derive clinically relevant inferences about study populations.
Semantically-aware population health risk analyses
Nov 27, 2018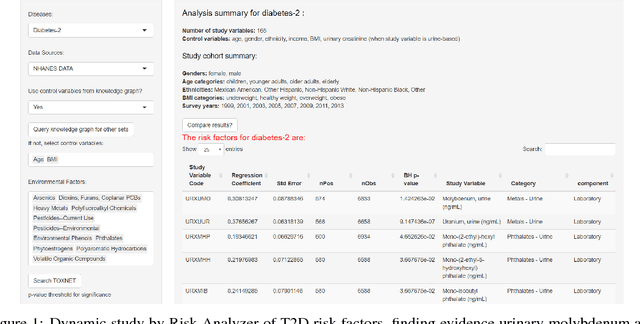

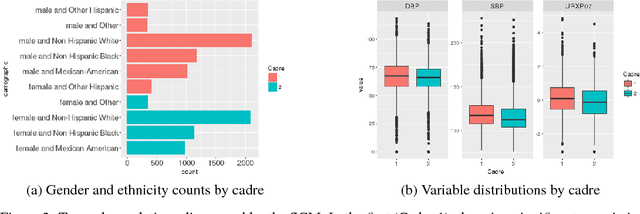
Abstract:One primary task of population health analysis is the identification of risk factors that, for some subpopulation, have a significant association with some health condition. Examples include finding lifestyle factors associated with chronic diseases and finding genetic mutations associated with diseases in precision health. We develop a combined semantic and machine learning system that uses a health risk ontology and knowledge graph (KG) to dynamically discover risk factors and their associated subpopulations. Semantics and the novel supervised cadre model make our system explainable. Future population health studies are easily performed and documented with provenance by specifying additional input and output KG cartridges.
Cadre Modeling: Simultaneously Discovering Subpopulations and Predictive Models
Oct 23, 2018



Abstract:We consider the problem in regression analysis of identifying subpopulations that exhibit different patterns of response, where each subpopulation requires a different underlying model. Unlike statistical cohorts, these subpopulations are not known a priori; thus, we refer to them as cadres. When the cadres and their associated models are interpretable, modeling leads to insights about the subpopulations and their associations with the regression target. We introduce a discriminative model that simultaneously learns cadre assignment and target-prediction rules. Sparsity-inducing priors are placed on the model parameters, under which independent feature selection is performed for both the cadre assignment and target-prediction processes. We learn models using adaptive step size stochastic gradient descent, and we assess cadre quality with bootstrapped sample analysis. We present simulated results showing that, when the true clustering rule does not depend on the entire set of features, our method significantly outperforms methods that learn subpopulation-discovery and target-prediction rules separately. In a materials-by-design case study, our model provides state-of-the-art prediction of polymer glass transition temperature. Importantly, the method identifies cadres of polymers that respond differently to structural perturbations, thus providing design insight for targeting or avoiding specific transition temperature ranges. It identifies chemically meaningful cadres, each with interpretable models. Further experimental results show that cadre methods have generalization that is competitive with linear and nonlinear regression models and can identify robust subpopulations.
* 8 pages, 6 figures
A Precision Environment-Wide Association Study of Hypertension via Supervised Cadre Models
Aug 14, 2018



Abstract:We consider the problem in precision health of grouping people into subpopulations based on their degree of vulnerability to a risk factor. These subpopulations cannot be discovered with traditional clustering techniques because their quality is evaluated with a supervised metric: the ease of modeling a response variable over observations within them. Instead, we apply the supervised cadre model (SCM), which does use this metric. We extend the SCM formalism so that it may be applied to multivariate regression and binary classification problems. We also develop a way to use conditional entropy to assess the confidence in the process by which a subject is assigned their cadre. Using the SCM, we generalize the environment-wide association study (EWAS) workflow to be able to model heterogeneity in population risk. In our EWAS, we consider more than two hundred environmental exposure factors and find their association with diastolic blood pressure, systolic blood pressure, and hypertension. This requires adapting the SCM to be applicable to data generated by a complex survey design. After correcting for false positives, we found 25 exposure variables that had a significant association with at least one of our response variables. Eight of these were significant for a discovered subpopulation but not for the overall population. Some of these associations have been identified by previous researchers, while others appear to be novel associations. We examine several learned subpopulations in detail, and we find that they are interpretable and that they suggest further research questions.
Knowledge Integration for Disease Characterization: A Breast Cancer Example
Jul 20, 2018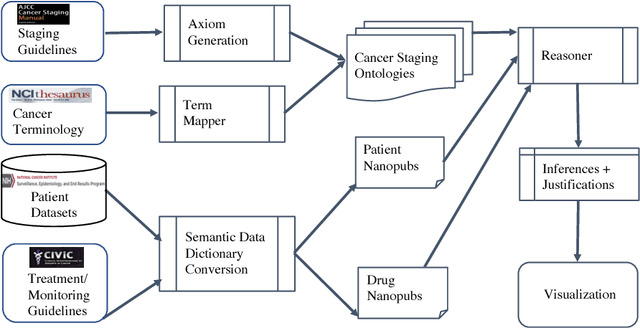

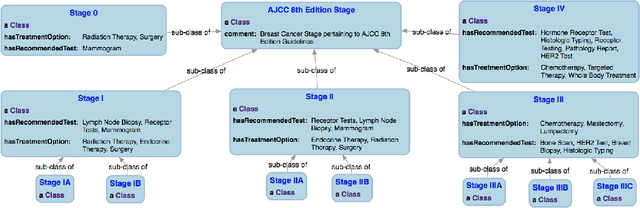
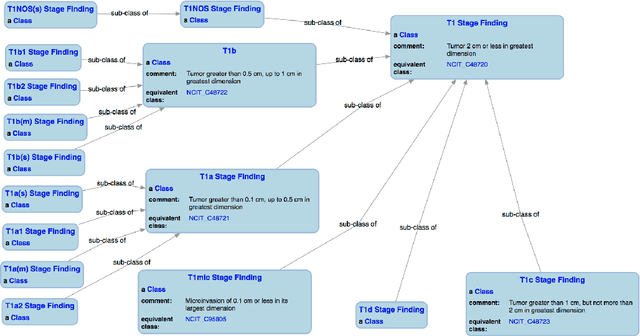
Abstract:With the rapid advancements in cancer research, the information that is useful for characterizing disease, staging tumors, and creating treatment and survivorship plans has been changing at a pace that creates challenges when physicians try to remain current. One example involves increasing usage of biomarkers when characterizing the pathologic prognostic stage of a breast tumor. We present our semantic technology approach to support cancer characterization and demonstrate it in our end-to-end prototype system that collects the newest breast cancer staging criteria from authoritative oncology manuals to construct an ontology for breast cancer. Using a tool we developed that utilizes this ontology, physician-facing applications can be used to quickly stage a new patient to support identifying risks, treatment options, and monitoring plans based on authoritative and best practice guidelines. Physicians can also re-stage existing patients or patient populations, allowing them to find patients whose stage has changed in a given patient cohort. As new guidelines emerge, using our proposed mechanism, which is grounded by semantic technologies for ingesting new data from staging manuals, we have created an enriched cancer staging ontology that integrates relevant data from several sources with very little human intervention.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge