Adrien Pavao
Winning solutions and post-challenge analyses of the ChaLearn AutoDL challenge 2019
Jan 11, 2022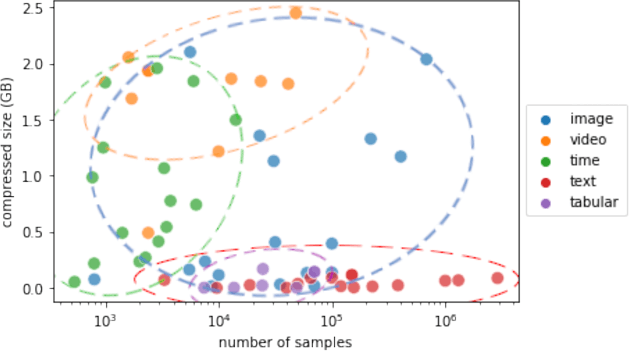
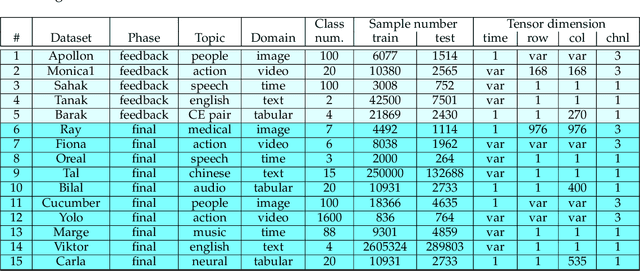
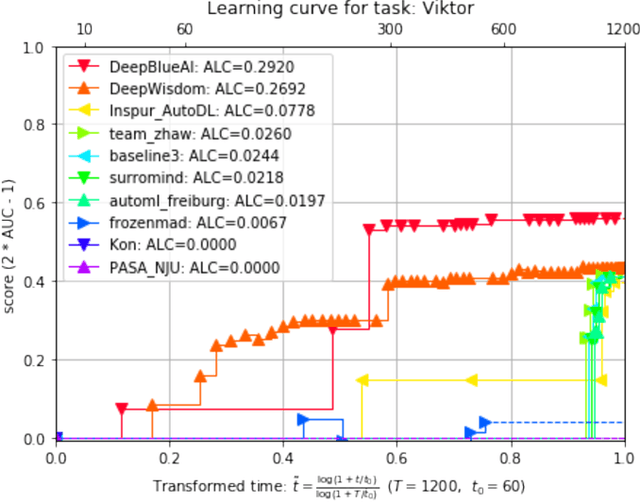
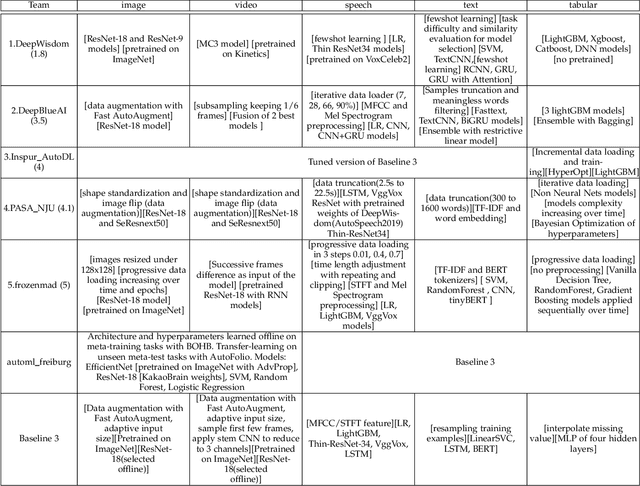
Abstract:This paper reports the results and post-challenge analyses of ChaLearn's AutoDL challenge series, which helped sorting out a profusion of AutoML solutions for Deep Learning (DL) that had been introduced in a variety of settings, but lacked fair comparisons. All input data modalities (time series, images, videos, text, tabular) were formatted as tensors and all tasks were multi-label classification problems. Code submissions were executed on hidden tasks, with limited time and computational resources, pushing solutions that get results quickly. In this setting, DL methods dominated, though popular Neural Architecture Search (NAS) was impractical. Solutions relied on fine-tuned pre-trained networks, with architectures matching data modality. Post-challenge tests did not reveal improvements beyond the imposed time limit. While no component is particularly original or novel, a high level modular organization emerged featuring a "meta-learner", "data ingestor", "model selector", "model/learner", and "evaluator". This modularity enabled ablation studies, which revealed the importance of (off-platform) meta-learning, ensembling, and efficient data management. Experiments on heterogeneous module combinations further confirm the (local) optimality of the winning solutions. Our challenge legacy includes an ever-lasting benchmark (http://autodl.chalearn.org), the open-sourced code of the winners, and a free "AutoDL self-service".
* The first three authors contributed equally; This is only a draft version
Synthetic Event Time Series Health Data Generation
Nov 27, 2019



Abstract:Synthetic medical data which preserves privacy while maintaining utility can be used as an alternative to real medical data, which has privacy costs and resource constraints associated with it. At present, most models focus on generating cross-sectional health data which is not necessarily representative of real data. In reality, medical data is longitudinal in nature, with a single patient having multiple health events, non-uniformly distributed throughout their lifetime. These events are influenced by patient covariates such as comorbidities, age group, gender etc. as well as external temporal effects (e.g. flu season). While there exist seminal methods to model time series data, it becomes increasingly challenging to extend these methods to medical event time series data. Due to the complexity of the real data, in which each patient visit is an event, we transform the data by using summary statistics to characterize the events for a fixed set of time intervals, to facilitate analysis and interpretability. We then train a generative adversarial network to generate synthetic data. We demonstrate this approach by generating human sleep patterns, from a publicly available dataset. We empirically evaluate the generated data and show close univariate resemblance between synthetic and real data. However, we also demonstrate how stratification by covariates is required to gain a deeper understanding of synthetic data quality.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge