James P. McCusker
Geospatial Reasoning with Shapefiles for Supporting Policy Decisions
Jun 09, 2021

Abstract:Policies are authoritative assets that are present in multiple domains to support decision-making. They describe what actions are allowed or recommended when domain entities and their attributes satisfy certain criteria. It is common to find policies that contain geographical rules, including distance and containment relationships among named locations. These locations' polygons can often be found encoded in geospatial datasets. We present an approach to transform data from geospatial datasets into Linked Data using the OWL, PROV-O, and GeoSPARQL standards, and to leverage this representation to support automated ontology-based policy decisions. We applied our approach to location-sensitive radio spectrum policies to identify relationships between radio transmitters coordinates and policy-regulated regions in Census.gov datasets. Using a policy evaluation pipeline that mixes OWL reasoning and GeoSPARQL, our approach implements the relevant geospatial relationships, according to a set of requirements elicited by radio spectrum domain experts.
Graph4Code: A Machine Interpretable Knowledge Graph for Code
Feb 21, 2020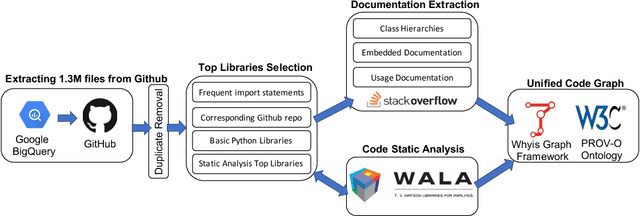
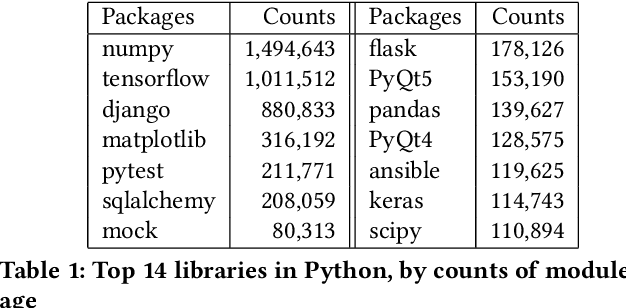
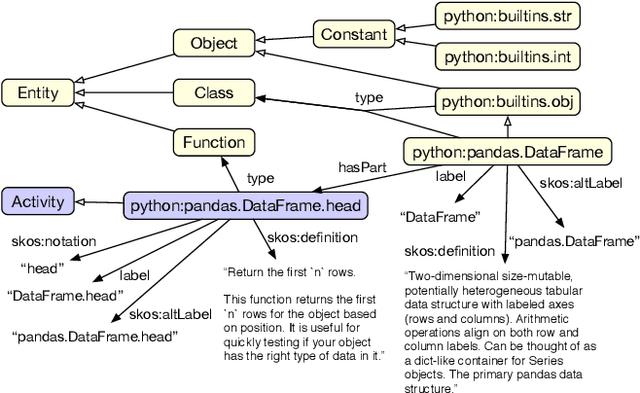

Abstract:Knowledge graphs have proven to be extremely useful in powering diverse applications in semantic search, natural language understanding, and even image classification. Graph4Code attempts to build well structured knowledge graphs about program code to similarly revolutionize diverse applications such as code search, code understanding, refactoring, bug detection, and code automation. We build such a graph by applying a set of generic code analysis techniques to Python code on the web. Since use of popular Python modules is ubiquitous in code, calls to functions in Python modules serve as key nodes of the knowledge graph. The edges in the graph are based on 1) function usage in the wild (e.g., which other function tends to call this one, or which function tends to precede this one, as gleaned from program analysis), 2) documentation about the function (e.g., code documentation, usage documentation, or forum discussions such as StackOverflow), and 3) program specific features such as class hierarchies. We use the Whyis knowledge graph management framework to make the graph easily extensible. We apply these techniques to 1.3M Python files drawn from GitHub, and associated documentation on the web for over 400 popular libraries, as well as StackOverflow posts about the same set of libraries. This knowledge graph will be made available soon to the larger community for use.
Making Study Populations Visible through Knowledge Graphs
Jul 09, 2019



Abstract:Treatment recommendations within Clinical Practice Guidelines (CPGs) are largely based on findings from clinical trials and case studies, referred to here as research studies, that are often based on highly selective clinical populations, referred to here as study cohorts. When medical practitioners apply CPG recommendations, they need to understand how well their patient population matches the characteristics of those in the study cohort, and thus are confronted with the challenges of locating the study cohort information and making an analytic comparison. To address these challenges, we develop an ontology-enabled prototype system, which exposes the population descriptions in research studies in a declarative manner, with the ultimate goal of allowing medical practitioners to better understand the applicability and generalizability of treatment recommendations. We build a Study Cohort Ontology (SCO) to encode the vocabulary of study population descriptions, that are often reported in the first table in the published work, thus they are often referred to as Table 1. We leverage the well-used Semanticscience Integrated Ontology (SIO) for defining property associations between classes. Further, we model the key components of Table 1s, i.e., collections of study subjects, subject characteristics, and statistical measures in RDF knowledge graphs. We design scenarios for medical practitioners to perform population analysis, and generate cohort similarity visualizations to determine the applicability of a study population to the clinical population of interest. Our semantic approach to make study populations visible, by standardized representations of Table 1s, allows users to quickly derive clinically relevant inferences about study populations.
Knowledge Integration for Disease Characterization: A Breast Cancer Example
Jul 20, 2018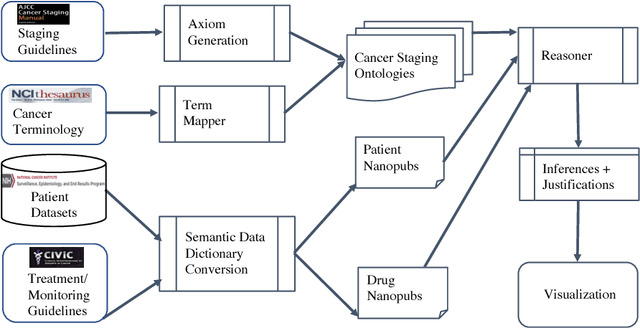

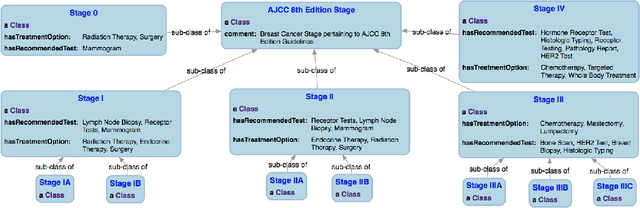
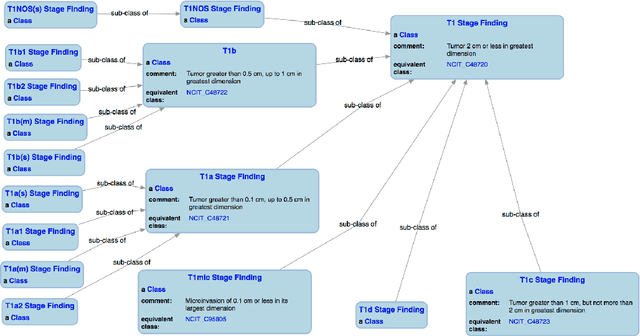
Abstract:With the rapid advancements in cancer research, the information that is useful for characterizing disease, staging tumors, and creating treatment and survivorship plans has been changing at a pace that creates challenges when physicians try to remain current. One example involves increasing usage of biomarkers when characterizing the pathologic prognostic stage of a breast tumor. We present our semantic technology approach to support cancer characterization and demonstrate it in our end-to-end prototype system that collects the newest breast cancer staging criteria from authoritative oncology manuals to construct an ontology for breast cancer. Using a tool we developed that utilizes this ontology, physician-facing applications can be used to quickly stage a new patient to support identifying risks, treatment options, and monitoring plans based on authoritative and best practice guidelines. Physicians can also re-stage existing patients or patient populations, allowing them to find patients whose stage has changed in a given patient cohort. As new guidelines emerge, using our proposed mechanism, which is grounded by semantic technologies for ingesting new data from staging manuals, we have created an enriched cancer staging ontology that integrates relevant data from several sources with very little human intervention.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge