Anthony Philippakis
Prediction-powered Generalization of Causal Inferences
Jun 05, 2024Abstract:Causal inferences from a randomized controlled trial (RCT) may not pertain to a target population where some effect modifiers have a different distribution. Prior work studies generalizing the results of a trial to a target population with no outcome but covariate data available. We show how the limited size of trials makes generalization a statistically infeasible task, as it requires estimating complex nuisance functions. We develop generalization algorithms that supplement the trial data with a prediction model learned from an additional observational study (OS), without making any assumptions on the OS. We theoretically and empirically show that our methods facilitate better generalization when the OS is high-quality, and remain robust when it is not, and e.g., have unmeasured confounding.
Non-Invasive Medical Digital Twins using Physics-Informed Self-Supervised Learning
Feb 29, 2024Abstract:A digital twin is a virtual replica of a real-world physical phenomena that uses mathematical modeling to characterize and simulate its defining features. By constructing digital twins for disease processes, we can perform in-silico simulations that mimic patients' health conditions and counterfactual outcomes under hypothetical interventions in a virtual setting. This eliminates the need for invasive procedures or uncertain treatment decisions. In this paper, we propose a method to identify digital twin model parameters using only noninvasive patient health data. We approach the digital twin modeling as a composite inverse problem, and observe that its structure resembles pretraining and finetuning in self-supervised learning (SSL). Leveraging this, we introduce a physics-informed SSL algorithm that initially pretrains a neural network on the pretext task of solving the physical model equations. Subsequently, the model is trained to reconstruct low-dimensional health measurements from noninvasive modalities while being constrained by the physical equations learned in pretraining. We apply our method to identify digital twins of cardiac hemodynamics using noninvasive echocardiogram videos, and demonstrate its utility in unsupervised disease detection and in-silico clinical trials.
Benchmarking Observational Studies with Experimental Data under Right-Censoring
Feb 23, 2024


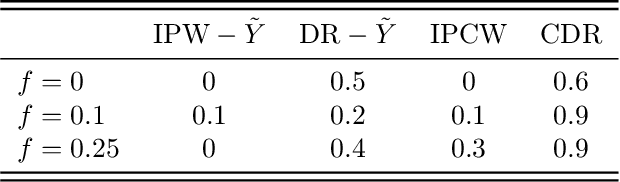
Abstract:Drawing causal inferences from observational studies (OS) requires unverifiable validity assumptions; however, one can falsify those assumptions by benchmarking the OS with experimental data from a randomized controlled trial (RCT). A major limitation of existing procedures is not accounting for censoring, despite the abundance of RCTs and OSes that report right-censored time-to-event outcomes. We consider two cases where censoring time (1) is independent of time-to-event and (2) depends on time-to-event the same way in OS and RCT. For the former, we adopt a censoring-doubly-robust signal for the conditional average treatment effect (CATE) to facilitate an equivalence test of CATEs in OS and RCT, which serves as a proxy for testing if the validity assumptions hold. For the latter, we show that the same test can still be used even though unbiased CATE estimation may not be possible. We verify the effectiveness of our censoring-aware tests via semi-synthetic experiments and analyze RCT and OS data from the Women's Health Initiative study.
Latent Space Explorer: Visual Analytics for Multimodal Latent Space Exploration
Dec 01, 2023Abstract:Machine learning models built on training data with multiple modalities can reveal new insights that are not accessible through unimodal datasets. For example, cardiac magnetic resonance images (MRIs) and electrocardiograms (ECGs) are both known to capture useful information about subjects' cardiovascular health status. A multimodal machine learning model trained from large datasets can potentially predict the onset of heart-related diseases and provide novel medical insights about the cardiovascular system. Despite the potential benefits, it is difficult for medical experts to explore multimodal representation models without visual aids and to test the predictive performance of the models on various subpopulations. To address the challenges, we developed a visual analytics system called Latent Space Explorer. Latent Space Explorer provides interactive visualizations that enable users to explore the multimodal representation of subjects, define subgroups of interest, interactively decode data with different modalities with the selected subjects, and inspect the accuracy of the embedding in downstream prediction tasks. A user study was conducted with medical experts and their feedback provided useful insights into how Latent Space Explorer can help their analysis and possible new direction for further development in the medical domain.
Generating Drug Repurposing Hypotheses through the Combination of Disease-Specific Hypergraphs
Nov 16, 2023


Abstract:The drug development pipeline for a new compound can last 10-20 years and cost over 10 billion. Drug repurposing offers a more time- and cost-effective alternative. Computational approaches based on biomedical knowledge graph representations have recently yielded new drug repurposing hypotheses. In this study, we present a novel, disease-specific hypergraph representation learning technique to derive contextual embeddings of biological pathways of various lengths but that all start at any given drug and all end at the disease of interest. Further, we extend this method to multi-disease hypergraphs. To determine the repurposing potential of each of the 1,522 drugs, we derive drug-specific distributions of cosine similarity values and ultimately consider the median for ranking. Cosine similarity values are computed between (1) all biological pathways starting at the considered drug and ending at the disease of interest and (2) all biological pathways starting at drugs currently prescribed against that disease and ending at the disease of interest. We illustrate our approach with Alzheimer's disease (AD) and two of its risk factors: hypertension (HTN) and type 2 diabetes (T2D). We compare each drug's rank across four hypergraph settings (single- or multi-disease): AD only, AD + HTN, AD + T2D, and AD + HTN + T2D. Notably, our framework led to the identification of two promising drugs whose repurposing potential was significantly higher in hypergraphs combining two diseases: dapagliflozin (antidiabetic; moved up, from top 32$\%$ to top 7$\%$, across all considered drugs) and debrisoquine (antihypertensive; moved up, from top 76$\%$ to top 23$\%$). Our approach serves as a hypothesis generation tool, to be paired with a validation pipeline relying on laboratory experiments and semi-automated parsing of the biomedical literature.
InstructCV: Instruction-Tuned Text-to-Image Diffusion Models as Vision Generalists
Sep 30, 2023



Abstract:Recent advances in generative diffusion models have enabled text-controlled synthesis of realistic and diverse images with impressive quality. Despite these remarkable advances, the application of text-to-image generative models in computer vision for standard visual recognition tasks remains limited. The current de facto approach for these tasks is to design model architectures and loss functions that are tailored to the task at hand. In this paper, we develop a unified language interface for computer vision tasks that abstracts away task-specific design choices and enables task execution by following natural language instructions. Our approach involves casting multiple computer vision tasks as text-to-image generation problems. Here, the text represents an instruction describing the task, and the resulting image is a visually-encoded task output. To train our model, we pool commonly-used computer vision datasets covering a range of tasks, including segmentation, object detection, depth estimation, and classification. We then use a large language model to paraphrase prompt templates that convey the specific tasks to be conducted on each image, and through this process, we create a multi-modal and multi-task training dataset comprising input and output images along with annotated instructions. Following the InstructPix2Pix architecture, we apply instruction-tuning to a text-to-image diffusion model using our constructed dataset, steering its functionality from a generative model to an instruction-guided multi-task vision learner. Experiments demonstrate that our model, dubbed InstructCV, performs competitively compared to other generalist and task-specific vision models. Moreover, it exhibits compelling generalization capabilities to unseen data, categories, and user instructions.
Signature Activation: A Sparse Signal View for Holistic Saliency
Sep 20, 2023Abstract:The adoption of machine learning in healthcare calls for model transparency and explainability. In this work, we introduce Signature Activation, a saliency method that generates holistic and class-agnostic explanations for Convolutional Neural Network (CNN) outputs. Our method exploits the fact that certain kinds of medical images, such as angiograms, have clear foreground and background objects. We give theoretical explanation to justify our methods. We show the potential use of our method in clinical settings through evaluating its efficacy for aiding the detection of lesions in coronary angiograms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge