Irene Y. Chen
Uncertainty Drives Social Bias Changes in Quantized Large Language Models
Feb 05, 2026Abstract:Post-training quantization reduces the computational cost of large language models but fundamentally alters their social biases in ways that aggregate metrics fail to capture. We present the first large-scale study of 50 quantized models evaluated on PostTrainingBiasBench, a unified benchmark of 13 closed- and open-ended bias datasets. We identify a phenomenon we term quantization-induced masked bias flipping, in which up to 21% of responses flip between biased and unbiased states after quantization, despite showing no change in aggregate bias scores. These flips are strongly driven by model uncertainty, where the responses with high uncertainty are 3-11x more likely to change than the confident ones. Quantization strength amplifies this effect, with 4-bit quantized models exhibiting 4-6x more behavioral changes than 8-bit quantized models. Critically, these changes create asymmetric impacts across demographic groups, where bias can worsen by up to 18.6% for some groups while improving by 14.1% for others, yielding misleadingly neutral aggregate outcomes. Larger models show no consistent robustness advantage, and group-specific shifts vary unpredictably across model families. Our findings demonstrate that compression fundamentally alters bias patterns, requiring crucial post-quantization evaluation and interventions to ensure reliability in practice.
Do Sparse Autoencoders Identify Reasoning Features in Language Models?
Jan 09, 2026Abstract:We investigate whether sparse autoencoders (SAEs) identify genuine reasoning features in large language models (LLMs). Starting from features selected using standard contrastive activation methods, we introduce a falsification-oriented framework that combines causal token injection experiments and LLM-guided falsification to test whether feature activation reflects reasoning processes or superficial linguistic correlates. Across 20 configurations spanning multiple model families, layers, and reasoning datasets, we find that identified reasoning features are highly sensitive to token-level interventions. Injecting a small number of feature-associated tokens into non-reasoning text is sufficient to elicit strong activation for 59% to 94% of features, indicating reliance on lexical artifacts. For the remaining features that are not explained by simple token triggers, LLM-guided falsification consistently produces non-reasoning inputs that activate the feature and reasoning inputs that do not, with no analyzed feature satisfying our criteria for genuine reasoning behavior. Steering these features yields minimal changes or slight degradations in benchmark performance. Together, these results suggest that SAE features identified by contrastive approaches primarily capture linguistic correlates of reasoning rather than the underlying reasoning computations themselves.
Treatment Non-Adherence Bias in Clinical Machine Learning: A Real-World Study on Hypertension Medication
Feb 26, 2025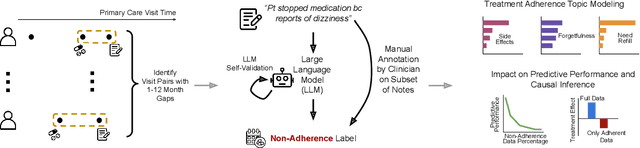
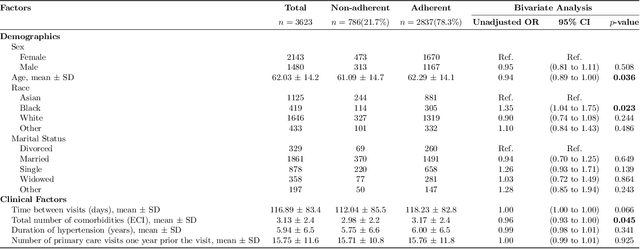
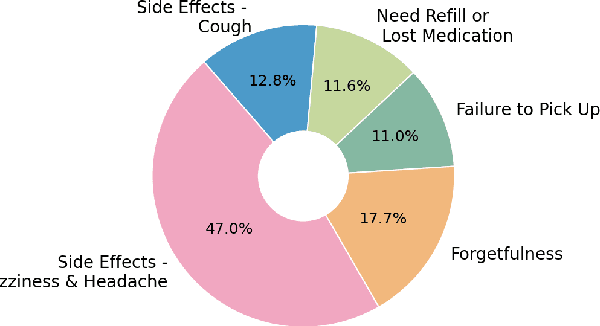
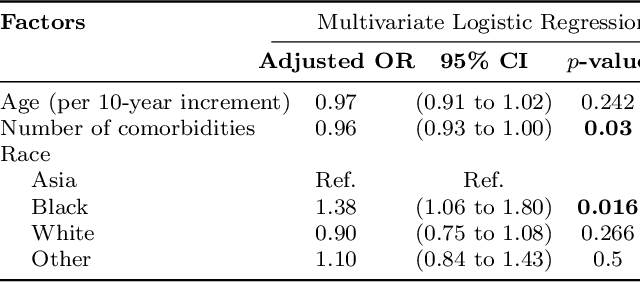
Abstract:Machine learning systems trained on electronic health records (EHRs) increasingly guide treatment decisions, but their reliability depends on the critical assumption that patients follow the prescribed treatments recorded in EHRs. Using EHR data from 3,623 hypertension patients, we investigate how treatment non-adherence introduces implicit bias that can fundamentally distort both causal inference and predictive modeling. By extracting patient adherence information from clinical notes using a large language model, we identify 786 patients (21.7%) with medication non-adherence. We further uncover key demographic and clinical factors associated with non-adherence, as well as patient-reported reasons including side effects and difficulties obtaining refills. Our findings demonstrate that this implicit bias can not only reverse estimated treatment effects, but also degrade model performance by up to 5% while disproportionately affecting vulnerable populations by exacerbating disparities in decision outcomes and model error rates. This highlights the importance of accounting for treatment non-adherence in developing responsible and equitable clinical machine learning systems.
Enhancing Semi-supervised Learning with Noisy Zero-shot Pseudolabels
Feb 18, 2025



Abstract:Semi-supervised learning (SSL) leverages limited labeled data alongside abundant unlabeled data to address labeling costs in machine learning. While recent foundation models enable zero-shot inference, attempts to integrate these capabilities into SSL through pseudo-labeling have shown mixed results due to unreliable zero-shot predictions. We present ZMT (Zero-Shot Multi-Task Learning), a framework that jointly optimizes zero-shot pseudo-labels and unsupervised representation learning objectives from contemporary SSL approaches. Our method introduces a multi-task learning-based mechanism that incorporates pseudo-labels while ensuring robustness to varying pseudo-label quality. Experiments across 8 datasets in vision, language, and audio domains demonstrate that ZMT reduces error by up to 56% compared to traditional SSL methods, with particularly compelling results when pseudo-labels are noisy and unreliable. ZMT represents a significant step toward making semi-supervised learning more effective and accessible in resource-constrained environments.
Deep Speech Synthesis from Multimodal Articulatory Representations
Dec 17, 2024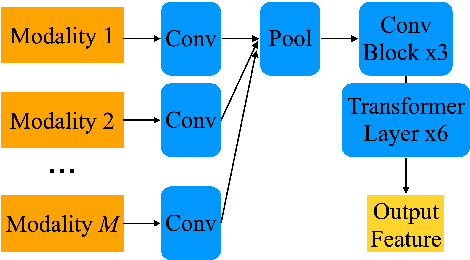
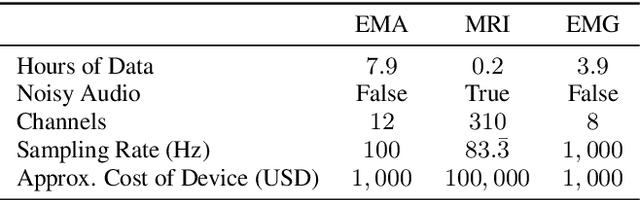
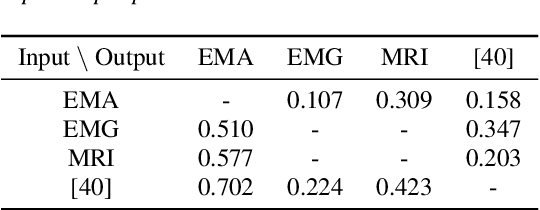
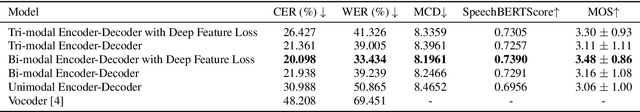
Abstract:The amount of articulatory data available for training deep learning models is much less compared to acoustic speech data. In order to improve articulatory-to-acoustic synthesis performance in these low-resource settings, we propose a multimodal pre-training framework. On single-speaker speech synthesis tasks from real-time magnetic resonance imaging and surface electromyography inputs, the intelligibility of synthesized outputs improves noticeably. For example, compared to prior work, utilizing our proposed transfer learning methods improves the MRI-to-speech performance by 36% word error rate. In addition to these intelligibility results, our multimodal pre-trained models consistently outperform unimodal baselines on three objective and subjective synthesis quality metrics.
Generative AI in Medicine
Dec 13, 2024

Abstract:The increased capabilities of generative AI have dramatically expanded its possible use cases in medicine. We provide a comprehensive overview of generative AI use cases for clinicians, patients, clinical trial organizers, researchers, and trainees. We then discuss the many challenges -- including maintaining privacy and security, improving transparency and interpretability, upholding equity, and rigorously evaluating models -- which must be overcome to realize this potential, and the open research directions they give rise to.
The Data Addition Dilemma
Aug 08, 2024Abstract:In many machine learning for healthcare tasks, standard datasets are constructed by amassing data across many, often fundamentally dissimilar, sources. But when does adding more data help, and when does it hinder progress on desired model outcomes in real-world settings? We identify this situation as the \textit{Data Addition Dilemma}, demonstrating that adding training data in this multi-source scaling context can at times result in reduced overall accuracy, uncertain fairness outcomes, and reduced worst-subgroup performance. We find that this possibly arises from an empirically observed trade-off between model performance improvements due to data scaling and model deterioration from distribution shift. We thus establish baseline strategies for navigating this dilemma, introducing distribution shift heuristics to guide decision-making on which data sources to add in data scaling, in order to yield the expected model performance improvements. We conclude with a discussion of the required considerations for data collection and suggestions for studying data composition and scale in the age of increasingly larger models.
Updating the Minimum Information about CLinical Artificial Intelligence (MI-CLAIM) checklist for generative modeling research
Mar 05, 2024Abstract:Recent advances in generative models, including large language models (LLMs), vision language models (VLMs), and diffusion models, have accelerated the field of natural language and image processing in medicine and marked a significant paradigm shift in how biomedical models can be developed and deployed. While these models are highly adaptable to new tasks, scaling and evaluating their usage presents new challenges not addressed in previous frameworks. In particular, the ability of these models to produce useful outputs with little to no specialized training data ("zero-" or "few-shot" approaches), as well as the open-ended nature of their outputs, necessitate the development of updated guidelines in using and evaluating these models. In response to gaps in standards and best practices for the development of clinical AI tools identified by US Executive Order 141103 and several emerging national networks for clinical AI evaluation, we begin to formalize some of these guidelines by building on the "Minimum information about clinical artificial intelligence modeling" (MI-CLAIM) checklist. The MI-CLAIM checklist, originally developed in 2020, provided a set of six steps with guidelines on the minimum information necessary to encourage transparent, reproducible research for artificial intelligence (AI) in medicine. Here, we propose modifications to the original checklist that highlight differences in training, evaluation, interpretability, and reproducibility of generative models compared to traditional AI models for clinical research. This updated checklist also seeks to clarify cohort selection reporting and adds additional items on alignment with ethical standards.
Identifying Reasons for Contraceptive Switching from Real-World Data Using Large Language Models
Feb 06, 2024Abstract:Prescription contraceptives play a critical role in supporting women's reproductive health. With nearly 50 million women in the United States using contraceptives, understanding the factors that drive contraceptives selection and switching is of significant interest. However, many factors related to medication switching are often only captured in unstructured clinical notes and can be difficult to extract. Here, we evaluate the zero-shot abilities of a recently developed large language model, GPT-4 (via HIPAA-compliant Microsoft Azure API), to identify reasons for switching between classes of contraceptives from the UCSF Information Commons clinical notes dataset. We demonstrate that GPT-4 can accurately extract reasons for contraceptive switching, outperforming baseline BERT-based models with microF1 scores of 0.849 and 0.881 for contraceptive start and stop extraction, respectively. Human evaluation of GPT-4-extracted reasons for switching showed 91.4% accuracy, with minimal hallucinations. Using extracted reasons, we identified patient preference, adverse events, and insurance as key reasons for switching using unsupervised topic modeling approaches. Notably, we also showed using our approach that "weight gain/mood change" and "insurance coverage" are disproportionately found as reasons for contraceptive switching in specific demographic populations. Our code and supplemental data are available at https://github.com/BMiao10/contraceptive-switching.
Designing Guiding Principles for NLP for Healthcare: A Case Study of Maternal Health
Dec 19, 2023



Abstract:Objective: An ethical framework for the use of large language models (LLMs) is urgently needed to shape how natural language processing (NLP) tools are used for healthcare applications. Drawing directly from the voices of those most affected, we propose a set of guiding principles for the use of NLP in healthcare, with examples based on applications in maternal health. Materials and Methods: We led an interactive session centered on an LLM-based chatbot demonstration during a full-day workshop with 39 participants, and additionally surveyed 30 healthcare workers and 30 birthing people about their values, needs, and perceptions of AI and LLMs. We conducted quantitative and qualitative analyses of the interactive discussions to consolidate our findings into a set of guiding principles. Results: Using the case study of maternal health, we propose nine principles for ethical use of LLMs, grouped into three categories: (i) contextual significance, (ii) measurements, and (iii) who/what is valued. We describe rationales underlying these principles and provide practical advice. Discussion: Healthcare faces existing challenges including the balance of power in clinician-patient relationships, systemic health disparities, historical injustices, and economic constraints. Our principles serve as a framework for surfacing key considerations when deploying LLMs in medicine, as well as providing a methodological pattern for other researchers to follow. Conclusion: This set of principles can serve as a resource to practitioners working on maternal health and other healthcare fields to emphasize the importance of technical nuance, historical context, and inclusive design when developing LLMs for use in clinical settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge