Ahmed Alaa
ReasonEdit: Editing Vision-Language Models using Human Reasoning
Feb 03, 2026Abstract:Model editing aims to correct errors in large, pretrained models without altering unrelated behaviors. While some recent works have edited vision-language models (VLMs), no existing editors tackle reasoning-heavy tasks, which typically require humans and models to reason about images. We therefore propose ReasonEdit, the first VLM editor to let users explain their reasoning during editing, introducing a new, practical model editing setup. ReasonEdit continuously stores human reasoning in a codebook, and retrieves only relevant facts during inference using a novel topology-balanced multimodal embedding method inspired by network science. Across four VLMs on multiple rationale-based visual question answering datasets, ReasonEdit achieves state-of-the-art editing performance, ultimately showing that using human reasoning during editing greatly improves edit generalization.
Aligning Language Model Benchmarks with Pairwise Preferences
Feb 02, 2026Abstract:Language model benchmarks are pervasive and computationally-efficient proxies for real-world performance. However, many recent works find that benchmarks often fail to predict real utility. Towards bridging this gap, we introduce benchmark alignment, where we use limited amounts of information about model performance to automatically update offline benchmarks, aiming to produce new static benchmarks that predict model pairwise preferences in given test settings. We then propose BenchAlign, the first solution to this problem, which learns preference-aligned weight- ings for benchmark questions using the question-level performance of language models alongside ranked pairs of models that could be collected during deployment, producing new benchmarks that rank previously unseen models according to these preferences. Our experiments show that our aligned benchmarks can accurately rank unseen models according to models of human preferences, even across different sizes, while remaining interpretable. Overall, our work provides insights into the limits of aligning benchmarks with practical human preferences, which stands to accelerate model development towards real utility.
Advances in LLM Reasoning Enable Flexibility in Clinical Problem-Solving
Jan 17, 2026Abstract:Large Language Models (LLMs) have achieved high accuracy on medical question-answer (QA) benchmarks, yet their capacity for flexible clinical reasoning has been debated. Here, we asked whether advances in reasoning LLMs improve their cognitive flexibility in clinical reasoning. We assessed reasoning models from the OpenAI, Grok, Gemini, Claude, and DeepSeek families on the medicine abstraction and reasoning corpus (mARC), an adversarial medical QA benchmark which utilizes the Einstellung effect to induce inflexible overreliance on learned heuristic patterns in contexts where they become suboptimal. We found that strong reasoning models avoided Einstellung-based traps more often than weaker reasoning models, achieving human-level performance on mARC. On questions most commonly missed by physicians, the top 5 performing models answered 55% to 70% correctly with high confidence, indicating that these models may be less susceptible than humans to Einstellung effects. Our results indicate that strong reasoning models demonstrate improved flexibility in medical reasoning, achieving performance on par with humans on mARC.
Data reuse enables cost-efficient randomized trials of medical AI models
Nov 13, 2025Abstract:Randomized controlled trials (RCTs) are indispensable for establishing the clinical value of medical artificial-intelligence (AI) tools, yet their high cost and long timelines hinder timely validation as new models emerge rapidly. Here, we propose BRIDGE, a data-reuse RCT design for AI-based risk models. AI risk models support a broad range of interventions, including screening, treatment selection, and clinical alerts. BRIDGE trials recycle participant-level data from completed trials of AI models when legacy and updated models make concordant predictions, thereby reducing the enrollment requirement for subsequent trials. We provide a practical checklist for investigators to assess whether reusing data from previous trials allows for valid causal inference and preserves type I error. Using real-world datasets across breast cancer, cardiovascular disease, and sepsis, we demonstrate concordance between successive AI models, with up to 64.8% overlap in top 5% high-risk cohorts. We then simulate a series of breast cancer screening studies, where our design reduced required enrollment by 46.6%--saving over US$2.8 million--while maintaining 80% power. By transforming trials into adaptive, modular studies, our proposed design makes Level I evidence generation feasible for every model iteration, thereby accelerating cost-effective translation of AI into routine care.
Hybrid Meta-learners for Estimating Heterogeneous Treatment Effects
Jun 16, 2025Abstract:Estimating conditional average treatment effects (CATE) from observational data involves modeling decisions that differ from supervised learning, particularly concerning how to regularize model complexity. Previous approaches can be grouped into two primary "meta-learner" paradigms that impose distinct inductive biases. Indirect meta-learners first fit and regularize separate potential outcome (PO) models and then estimate CATE by taking their difference, whereas direct meta-learners construct and directly regularize estimators for the CATE function itself. Neither approach consistently outperforms the other across all scenarios: indirect learners perform well when the PO functions are simple, while direct learners outperform when the CATE is simpler than individual PO functions. In this paper, we introduce the Hybrid Learner (H-learner), a novel regularization strategy that interpolates between the direct and indirect regularizations depending on the dataset at hand. The H-learner achieves this by learning intermediate functions whose difference closely approximates the CATE without necessarily requiring accurate individual approximations of the POs themselves. We demonstrate empirically that intentionally allowing suboptimal fits to the POs improves the bias-variance tradeoff in estimating CATE. Experiments conducted on semi-synthetic and real-world benchmark datasets illustrate that the H-learner consistently operates at the Pareto frontier, effectively combining the strengths of both direct and indirect meta-learners.
ER-REASON: A Benchmark Dataset for LLM-Based Clinical Reasoning in the Emergency Room
May 28, 2025



Abstract:Large language models (LLMs) have been extensively evaluated on medical question answering tasks based on licensing exams. However, real-world evaluations often depend on costly human annotators, and existing benchmarks tend to focus on isolated tasks that rarely capture the clinical reasoning or full workflow underlying medical decisions. In this paper, we introduce ER-Reason, a benchmark designed to evaluate LLM-based clinical reasoning and decision-making in the emergency room (ER)--a high-stakes setting where clinicians make rapid, consequential decisions across diverse patient presentations and medical specialties under time pressure. ER-Reason includes data from 3,984 patients, encompassing 25,174 de-identified longitudinal clinical notes spanning discharge summaries, progress notes, history and physical exams, consults, echocardiography reports, imaging notes, and ER provider documentation. The benchmark includes evaluation tasks that span key stages of the ER workflow: triage intake, initial assessment, treatment selection, disposition planning, and final diagnosis--each structured to reflect core clinical reasoning processes such as differential diagnosis via rule-out reasoning. We also collected 72 full physician-authored rationales explaining reasoning processes that mimic the teaching process used in residency training, and are typically absent from ER documentation. Evaluations of state-of-the-art LLMs on ER-Reason reveal a gap between LLM-generated and clinician-authored clinical reasoning for ER decisions, highlighting the need for future research to bridge this divide.
Model Editing with Graph-Based External Memory
May 23, 2025Abstract:Large language models (LLMs) have revolutionized natural language processing, yet their practical utility is often limited by persistent issues of hallucinations and outdated parametric knowledge. Although post-training model editing offers a pathway for dynamic updates, existing methods frequently suffer from overfitting and catastrophic forgetting. To tackle these challenges, we propose a novel framework that leverages hyperbolic geometry and graph neural networks for precise and stable model edits. We introduce HYPE (HYperbolic Parameter Editing), which comprises three key components: (i) Hyperbolic Graph Construction, which uses Poincar\'e embeddings to represent knowledge triples in hyperbolic space, preserving hierarchical relationships and preventing unintended side effects by ensuring that edits to parent concepts do not inadvertently affect child concepts; (ii) M\"obius-Transformed Updates, which apply hyperbolic addition to propagate edits while maintaining structural consistency within the hyperbolic manifold, unlike conventional Euclidean updates that distort relational distances; and (iii) Dual Stabilization, which combines gradient masking and periodic GNN parameter resetting to prevent catastrophic forgetting by focusing updates on critical parameters and preserving long-term knowledge. Experiments on CounterFact, CounterFact+, and MQuAKE with GPT-J and GPT2-XL demonstrate that HYPE significantly enhances edit stability, factual accuracy, and multi-hop reasoning.
Norm Growth and Stability Challenges in Localized Sequential Knowledge Editing
Feb 26, 2025Abstract:This study investigates the impact of localized updates to large language models (LLMs), specifically in the context of knowledge editing - a task aimed at incorporating or modifying specific facts without altering broader model capabilities. We first show that across different post-training interventions like continuous pre-training, full fine-tuning and LORA-based fine-tuning, the Frobenius norm of the updated matrices always increases. This increasing norm is especially detrimental for localized knowledge editing, where only a subset of matrices are updated in a model . We reveal a consistent phenomenon across various editing techniques, including fine-tuning, hypernetwork-based approaches, and locate-and-edit methods: the norm of the updated matrix invariably increases with successive updates. Such growth disrupts model balance, particularly when isolated matrices are updated while the rest of the model remains static, leading to potential instability and degradation of downstream performance. Upon deeper investigations of the intermediate activation vectors, we find that the norm of internal activations decreases and is accompanied by shifts in the subspaces occupied by these activations, which shows that these activation vectors now occupy completely different regions in the representation space compared to the unedited model. With our paper, we highlight the technical challenges with continuous and localized sequential knowledge editing and their implications for maintaining model stability and utility.
Generalized Venn and Venn-Abers Calibration with Applications in Conformal Prediction
Feb 08, 2025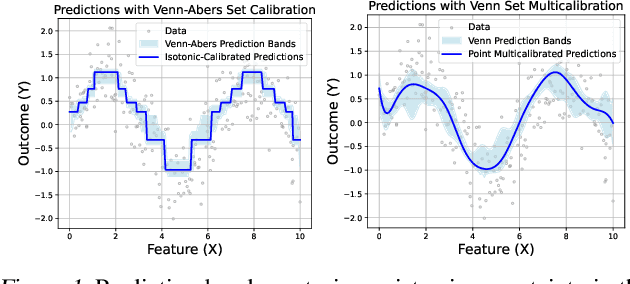
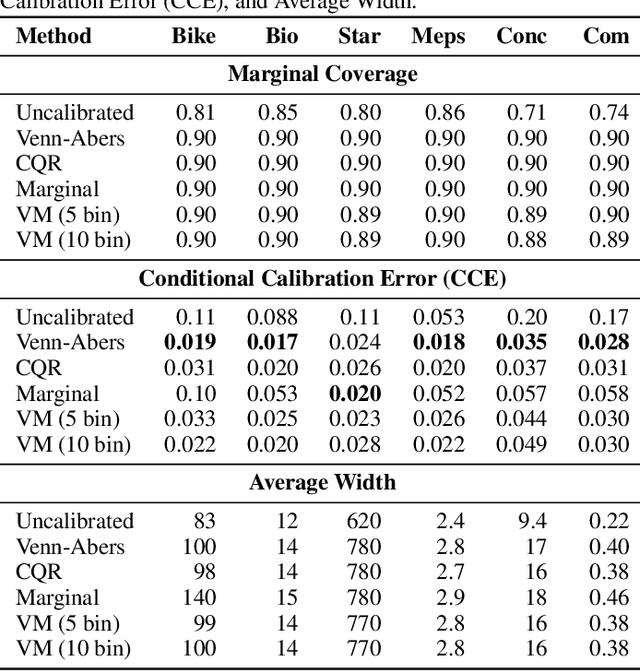
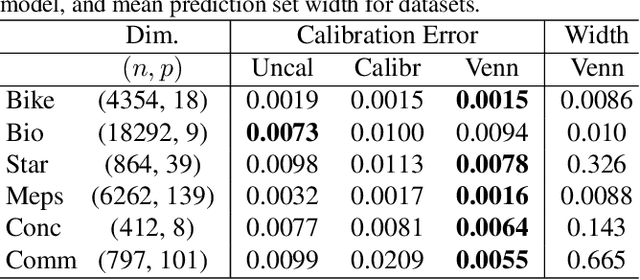
Abstract:Ensuring model calibration is critical for reliable predictions, yet popular distribution-free methods, such as histogram binning and isotonic regression, provide only asymptotic guarantees. We introduce a unified framework for Venn and Venn-Abers calibration, generalizing Vovk's binary classification approach to arbitrary prediction tasks and loss functions. Venn calibration leverages binning calibrators to construct prediction sets that contain at least one marginally perfectly calibrated point prediction in finite samples, capturing epistemic uncertainty in the calibration process. The width of these sets shrinks asymptotically to zero, converging to a conditionally calibrated point prediction. Furthermore, we propose Venn multicalibration, a novel methodology for finite-sample calibration across subpopulations. For quantile loss, group-conditional and multicalibrated conformal prediction arise as special cases of Venn multicalibration, and Venn calibration produces novel conformal prediction intervals that achieve quantile-conditional coverage. As a separate contribution, we extend distribution-free conditional calibration guarantees of histogram binning and isotonic calibration to general losses.
Limitations of Large Language Models in Clinical Problem-Solving Arising from Inflexible Reasoning
Feb 05, 2025Abstract:Large Language Models (LLMs) have attained human-level accuracy on medical question-answer (QA) benchmarks. However, their limitations in navigating open-ended clinical scenarios have recently been shown, raising concerns about the robustness and generalizability of LLM reasoning across diverse, real-world medical tasks. To probe potential LLM failure modes in clinical problem-solving, we present the medical abstraction and reasoning corpus (M-ARC). M-ARC assesses clinical reasoning through scenarios designed to exploit the Einstellung effect -- the fixation of thought arising from prior experience, targeting LLM inductive biases toward inflexible pattern matching from their training data rather than engaging in flexible reasoning. We find that LLMs, including current state-of-the-art o1 and Gemini models, perform poorly compared to physicians on M-ARC, often demonstrating lack of commonsense medical reasoning and a propensity to hallucinate. In addition, uncertainty estimation analyses indicate that LLMs exhibit overconfidence in their answers, despite their limited accuracy. The failure modes revealed by M-ARC in LLM medical reasoning underscore the need to exercise caution when deploying these models in clinical settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge