Amanda K. Hall
CancerGUIDE: Cancer Guideline Understanding via Internal Disagreement Estimation
Sep 09, 2025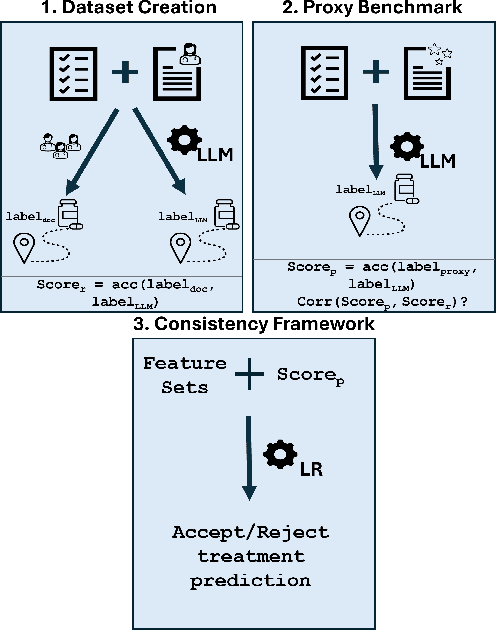
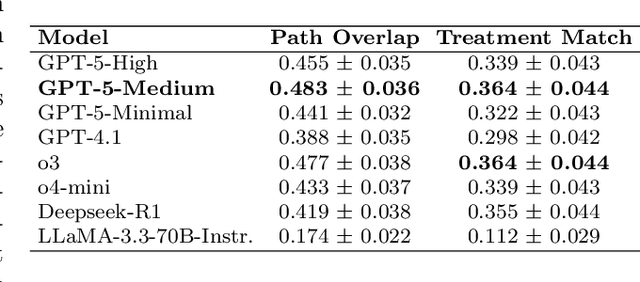
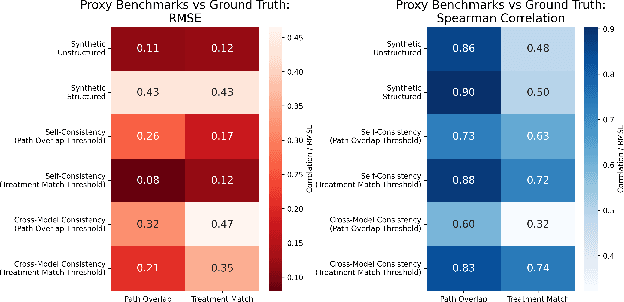
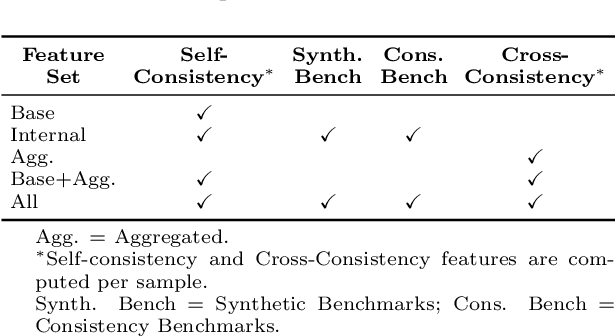
Abstract:The National Comprehensive Cancer Network (NCCN) provides evidence-based guidelines for cancer treatment. Translating complex patient presentations into guideline-compliant treatment recommendations is time-intensive, requires specialized expertise, and is prone to error. Advances in large language model (LLM) capabilities promise to reduce the time required to generate treatment recommendations and improve accuracy. We present an LLM agent-based approach to automatically generate guideline-concordant treatment trajectories for patients with non-small cell lung cancer (NSCLC). Our contributions are threefold. First, we construct a novel longitudinal dataset of 121 cases of NSCLC patients that includes clinical encounters, diagnostic results, and medical histories, each expertly annotated with the corresponding NCCN guideline trajectories by board-certified oncologists. Second, we demonstrate that existing LLMs possess domain-specific knowledge that enables high-quality proxy benchmark generation for both model development and evaluation, achieving strong correlation (Spearman coefficient r=0.88, RMSE = 0.08) with expert-annotated benchmarks. Third, we develop a hybrid approach combining expensive human annotations with model consistency information to create both the agent framework that predicts the relevant guidelines for a patient, as well as a meta-classifier that verifies prediction accuracy with calibrated confidence scores for treatment recommendations (AUROC=0.800), a critical capability for communicating the accuracy of outputs, custom-tailoring tradeoffs in performance, and supporting regulatory compliance. This work establishes a framework for clinically viable LLM-based guideline adherence systems that balance accuracy, interpretability, and regulatory requirements while reducing annotation costs, providing a scalable pathway toward automated clinical decision support.
BioAgents: Democratizing Bioinformatics Analysis with Multi-Agent Systems
Jan 10, 2025Abstract:Creating end-to-end bioinformatics workflows requires diverse domain expertise, which poses challenges for both junior and senior researchers as it demands a deep understanding of both genomics concepts and computational techniques. While large language models (LLMs) provide some assistance, they often fall short in providing the nuanced guidance needed to execute complex bioinformatics tasks, and require expensive computing resources to achieve high performance. We thus propose a multi-agent system built on small language models, fine-tuned on bioinformatics data, and enhanced with retrieval augmented generation (RAG). Our system, BioAgents, enables local operation and personalization using proprietary data. We observe performance comparable to human experts on conceptual genomics tasks, and suggest next steps to enhance code generation capabilities.
AI-Enhanced Sensemaking: Exploring the Design of a Generative AI-Based Assistant to Support Genetic Professionals
Dec 19, 2024



Abstract:Generative AI has the potential to transform knowledge work, but further research is needed to understand how knowledge workers envision using and interacting with generative AI. We investigate the development of generative AI tools to support domain experts in knowledge work, examining task delegation and the design of human-AI interactions. Our research focused on designing a generative AI assistant to aid genetic professionals in analyzing whole genome sequences (WGS) and other clinical data for rare disease diagnosis. Through interviews with 17 genetics professionals, we identified current challenges in WGS analysis. We then conducted co-design sessions with six genetics professionals to determine tasks that could be supported by an AI assistant and considerations for designing interactions with the AI assistant. From our findings, we identified sensemaking as both a current challenge in WGS analysis and a process that could be supported by AI. We contribute an understanding of how domain experts envision interacting with generative AI in their knowledge work, a detailed empirical study of WGS analysis, and three design considerations for using generative AI to support domain experts in sensemaking during knowledge work. CCS CONCEPTS: Human-centered computing, Human-computer interaction, Empirical studies in HCI Additional Keywords and Phrases: whole genome sequencing, generative AI, large language models, knowledge work, sensemaking, co-design, rare disease Contact Author: Angela Mastrianni (This work was done during the author's internship at Microsoft Research) Ashley Mae Conard and Amanda K. Hall contributed equally
ePillID Dataset: A Low-Shot Fine-Grained Benchmark for Pill Identification
May 28, 2020



Abstract:Identifying prescription medications is a frequent task for patients and medical professionals; however, this is an error-prone task as many pills have similar appearances (e.g. white round pills), which increases the risk of medication errors. In this paper, we introduce ePillID, the largest public benchmark on pill image recognition, composed of 13k images representing 8184 appearance classes (two sides for 4092 pill types). For most of the appearance classes, there exists only one reference image, making it a challenging low-shot recognition setting. We present our experimental setup and evaluation results of various baseline models on the benchmark. The best baseline using a multi-head metric-learning approach with bilinear features performed remarkably well; however, our error analysis suggests that they still fail to distinguish particularly confusing classes. The code and data are available at \url{https://github.com/usuyama/ePillID-benchmark}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge