Atul J. Butte
Dr-LLaVA: Visual Instruction Tuning with Symbolic Clinical Grounding
May 29, 2024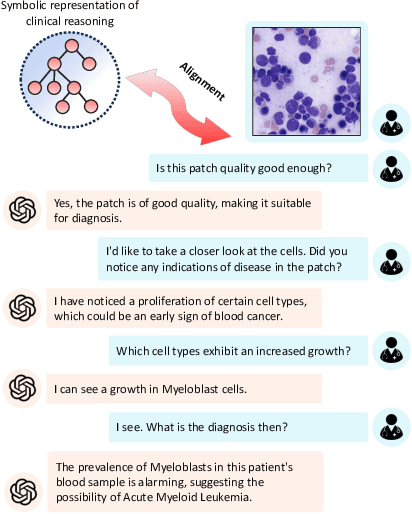
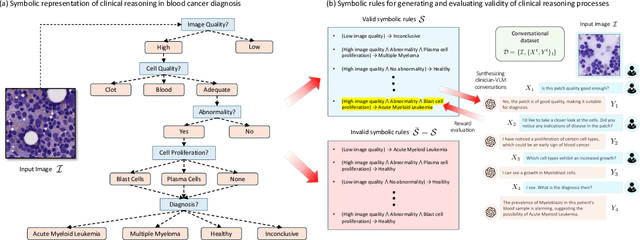
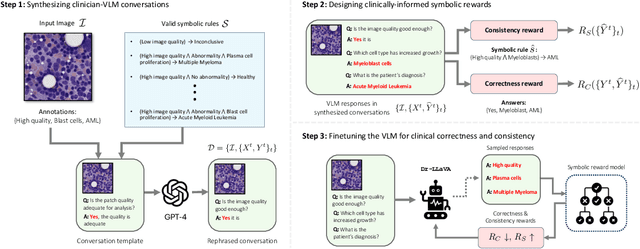
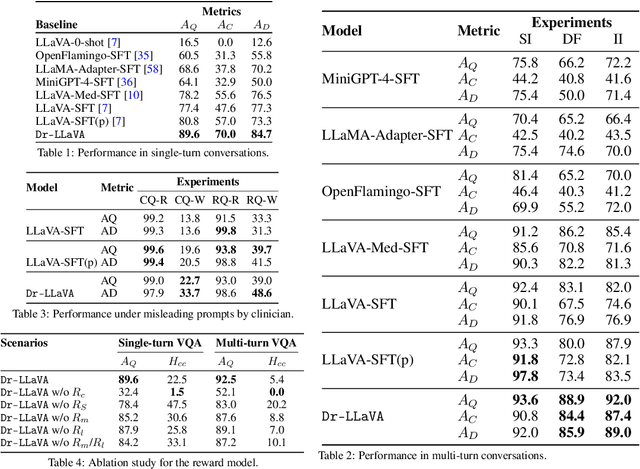
Abstract:Vision-Language Models (VLM) can support clinicians by analyzing medical images and engaging in natural language interactions to assist in diagnostic and treatment tasks. However, VLMs often exhibit "hallucinogenic" behavior, generating textual outputs not grounded in contextual multimodal information. This challenge is particularly pronounced in the medical domain, where we do not only require VLM outputs to be accurate in single interactions but also to be consistent with clinical reasoning and diagnostic pathways throughout multi-turn conversations. For this purpose, we propose a new alignment algorithm that uses symbolic representations of clinical reasoning to ground VLMs in medical knowledge. These representations are utilized to (i) generate GPT-4-guided visual instruction tuning data at scale, simulating clinician-VLM conversations with demonstrations of clinical reasoning, and (ii) create an automatic reward function that evaluates the clinical validity of VLM generations throughout clinician-VLM interactions. Our algorithm eliminates the need for human involvement in training data generation or reward model construction, reducing costs compared to standard reinforcement learning with human feedback (RLHF). We apply our alignment algorithm to develop Dr-LLaVA, a conversational VLM finetuned for analyzing bone marrow pathology slides, demonstrating strong performance in multi-turn medical conversations.
Updating the Minimum Information about CLinical Artificial Intelligence (MI-CLAIM) checklist for generative modeling research
Mar 05, 2024Abstract:Recent advances in generative models, including large language models (LLMs), vision language models (VLMs), and diffusion models, have accelerated the field of natural language and image processing in medicine and marked a significant paradigm shift in how biomedical models can be developed and deployed. While these models are highly adaptable to new tasks, scaling and evaluating their usage presents new challenges not addressed in previous frameworks. In particular, the ability of these models to produce useful outputs with little to no specialized training data ("zero-" or "few-shot" approaches), as well as the open-ended nature of their outputs, necessitate the development of updated guidelines in using and evaluating these models. In response to gaps in standards and best practices for the development of clinical AI tools identified by US Executive Order 141103 and several emerging national networks for clinical AI evaluation, we begin to formalize some of these guidelines by building on the "Minimum information about clinical artificial intelligence modeling" (MI-CLAIM) checklist. The MI-CLAIM checklist, originally developed in 2020, provided a set of six steps with guidelines on the minimum information necessary to encourage transparent, reproducible research for artificial intelligence (AI) in medicine. Here, we propose modifications to the original checklist that highlight differences in training, evaluation, interpretability, and reproducibility of generative models compared to traditional AI models for clinical research. This updated checklist also seeks to clarify cohort selection reporting and adds additional items on alignment with ethical standards.
Identifying Reasons for Contraceptive Switching from Real-World Data Using Large Language Models
Feb 06, 2024Abstract:Prescription contraceptives play a critical role in supporting women's reproductive health. With nearly 50 million women in the United States using contraceptives, understanding the factors that drive contraceptives selection and switching is of significant interest. However, many factors related to medication switching are often only captured in unstructured clinical notes and can be difficult to extract. Here, we evaluate the zero-shot abilities of a recently developed large language model, GPT-4 (via HIPAA-compliant Microsoft Azure API), to identify reasons for switching between classes of contraceptives from the UCSF Information Commons clinical notes dataset. We demonstrate that GPT-4 can accurately extract reasons for contraceptive switching, outperforming baseline BERT-based models with microF1 scores of 0.849 and 0.881 for contraceptive start and stop extraction, respectively. Human evaluation of GPT-4-extracted reasons for switching showed 91.4% accuracy, with minimal hallucinations. Using extracted reasons, we identified patient preference, adverse events, and insurance as key reasons for switching using unsupervised topic modeling approaches. Notably, we also showed using our approach that "weight gain/mood change" and "insurance coverage" are disproportionately found as reasons for contraceptive switching in specific demographic populations. Our code and supplemental data are available at https://github.com/BMiao10/contraceptive-switching.
A comparative study of zero-shot inference with large language models and supervised modeling in breast cancer pathology classification
Jan 25, 2024Abstract:Although supervised machine learning is popular for information extraction from clinical notes, creating large annotated datasets requires extensive domain expertise and is time-consuming. Meanwhile, large language models (LLMs) have demonstrated promising transfer learning capability. In this study, we explored whether recent LLMs can reduce the need for large-scale data annotations. We curated a manually-labeled dataset of 769 breast cancer pathology reports, labeled with 13 categories, to compare zero-shot classification capability of the GPT-4 model and the GPT-3.5 model with supervised classification performance of three model architectures: random forests classifier, long short-term memory networks with attention (LSTM-Att), and the UCSF-BERT model. Across all 13 tasks, the GPT-4 model performed either significantly better than or as well as the best supervised model, the LSTM-Att model (average macro F1 score of 0.83 vs. 0.75). On tasks with high imbalance between labels, the differences were more prominent. Frequent sources of GPT-4 errors included inferences from multiple samples and complex task design. On complex tasks where large annotated datasets cannot be easily collected, LLMs can reduce the burden of large-scale data labeling. However, if the use of LLMs is prohibitive, the use of simpler supervised models with large annotated datasets can provide comparable results. LLMs demonstrated the potential to speed up the execution of clinical NLP studies by reducing the need for curating large annotated datasets. This may result in an increase in the utilization of NLP-based variables and outcomes in observational clinical studies.
Extracting detailed oncologic history and treatment plan from medical oncology notes with large language models
Aug 07, 2023



Abstract:Both medical care and observational studies in oncology require a thorough understanding of a patient's disease progression and treatment history, often elaborately documented in clinical notes. Despite their vital role, no current oncology information representation and annotation schema fully encapsulates the diversity of information recorded within these notes. Although large language models (LLMs) have recently exhibited impressive performance on various medical natural language processing tasks, due to the current lack of comprehensively annotated oncology datasets, an extensive evaluation of LLMs in extracting and reasoning with the complex rhetoric in oncology notes remains understudied. We developed a detailed schema for annotating textual oncology information, encompassing patient characteristics, tumor characteristics, tests, treatments, and temporality. Using a corpus of 10 de-identified breast cancer progress notes at University of California, San Francisco, we applied this schema to assess the abilities of three recently-released LLMs (GPT-4, GPT-3.5-turbo, and FLAN-UL2) to perform zero-shot extraction of detailed oncological history from two narrative sections of clinical progress notes. Our team annotated 2750 entities, 2874 modifiers, and 1623 relationships. The GPT-4 model exhibited overall best performance, with an average BLEU score of 0.69, an average ROUGE score of 0.72, and an average accuracy of 67% on complex tasks (expert manual evaluation). Notably, it was proficient in tumor characteristic and medication extraction, and demonstrated superior performance in inferring symptoms due to cancer and considerations of future medications. The analysis demonstrates that GPT-4 is potentially already usable to extract important facts from cancer progress notes needed for clinical research, complex population management, and documenting quality patient care.
Revealing the impact of social circumstances on the selection of cancer therapy through natural language processing of social work notes
Jun 16, 2023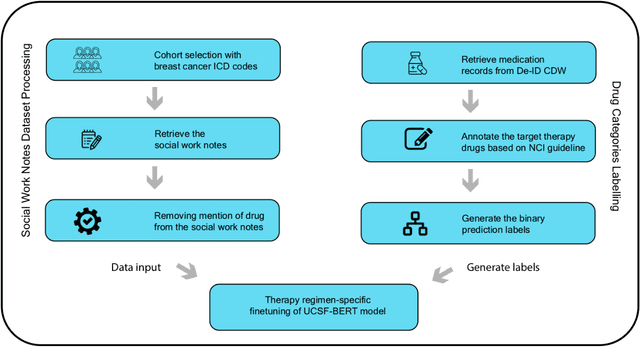
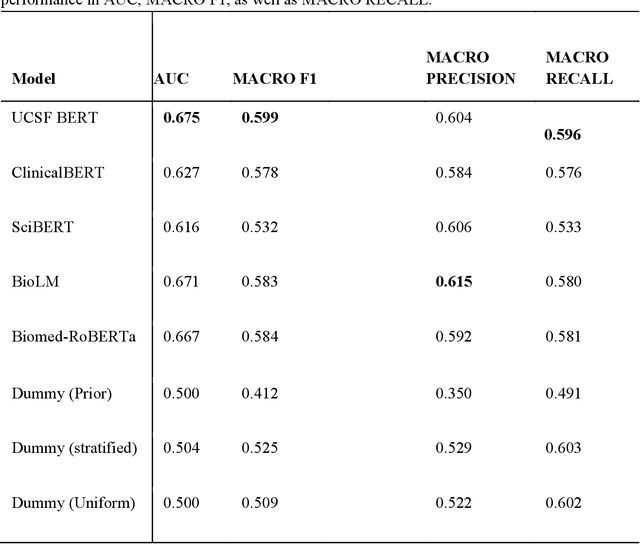
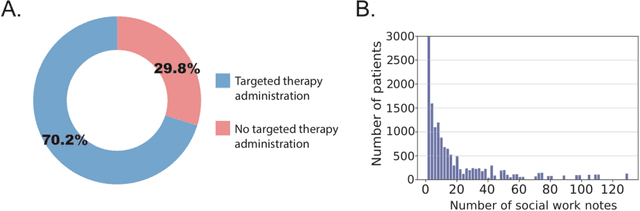
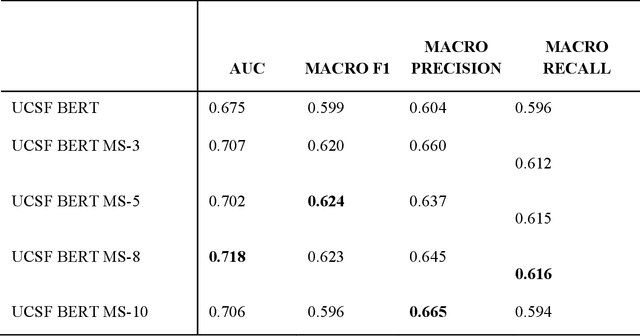
Abstract:We aimed to investigate the impact of social circumstances on cancer therapy selection using natural language processing to derive insights from social worker documentation. We developed and employed a Bidirectional Encoder Representations from Transformers (BERT) based approach, using a hierarchical multi-step BERT model (BERT-MS) to predict the prescription of targeted cancer therapy to patients based solely on documentation by clinical social workers. Our corpus included free-text clinical social work notes, combined with medication prescription information, for all patients treated for breast cancer. We conducted a feature importance analysis to pinpoint the specific social circumstances that impact cancer therapy selection. Using only social work notes, we consistently predicted the administration of targeted therapies, suggesting systematic differences in treatment selection exist due to non-clinical factors. The UCSF-BERT model, pretrained on clinical text at UCSF, outperformed other publicly available language models with an AUROC of 0.675 and a Macro F1 score of 0.599. The UCSF BERT-MS model, capable of leveraging multiple pieces of notes, surpassed the UCSF-BERT model in both AUROC and Macro-F1. Our feature importance analysis identified several clinically intuitive social determinants of health (SDOH) that potentially contribute to disparities in treatment. Our findings indicate that significant disparities exist among breast cancer patients receiving different types of therapies based on social determinants of health. Social work reports play a crucial role in understanding these disparities in clinical decision-making.
Cross-institution text mining to uncover clinical associations: a case study relating social factors and code status in intensive care medicine
Jan 16, 2023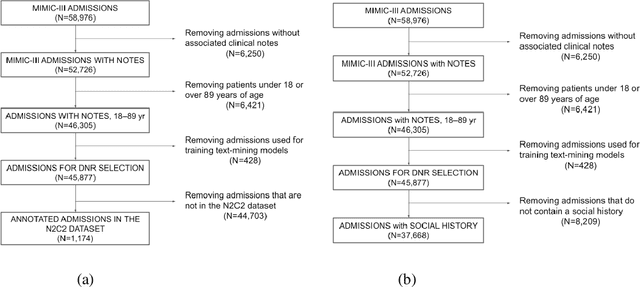

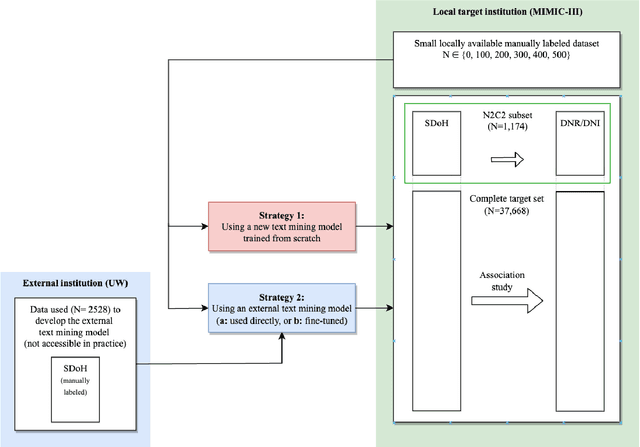
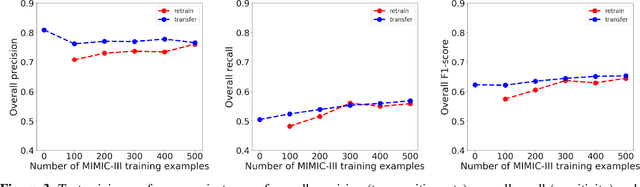
Abstract:Objective: Text mining of clinical notes embedded in electronic medical records is increasingly used to extract patient characteristics otherwise not or only partly available, to assess their association with relevant health outcomes. As manual data labeling needed to develop text mining models is resource intensive, we investigated whether off-the-shelf text mining models developed at external institutions, together with limited within-institution labeled data, could be used to reliably extract study variables to conduct association studies. Materials and Methods: We developed multiple text mining models on different combinations of within-institution and external-institution data to extract social factors from discharge reports of intensive care patients. Subsequently, we assessed the associations between social factors and having a do-not-resuscitate/intubate code. Results: Important differences were found between associations based on manually labeled data compared to text-mined social factors in three out of five cases. Adopting external-institution text mining models using manually labeled within-institution data resulted in models with higher F1-scores, but not in meaningfully different associations. Discussion: While text mining facilitated scaling analyses to larger samples leading to discovering a larger number of associations, the estimates may be unreliable. Confirmation is needed with better text mining models, ideally on a larger manually labeled dataset. Conclusion: The currently used text mining models were not sufficiently accurate to be used reliably in an association study. Model adaptation using within-institution data did not improve the estimates. Further research is needed to set conditions for reliable use of text mining in medical research.
Topic Modeling on Clinical Social Work Notes for Exploring Social Determinants of Health Factors
Dec 02, 2022Abstract:Most research studying social determinants of health (SDoH) has focused on physician notes or structured elements of the electronic medical record (EMR). We hypothesize that clinical notes from social workers, whose role is to ameliorate social and economic factors, might provide a richer source of data on SDoH. We sought to perform topic modeling to identify robust topics of discussion within a large cohort of social work notes. We retrieved a diverse, deidentified corpus of 0.95 million clinical social work notes from 181,644 patients at the University of California, San Francisco. We used word frequency analysis and Latent Dirichlet Allocation (LDA) topic modeling analysis to characterize this corpus and identify potential topics of discussion. Word frequency analysis identified both medical and non-medical terms associated with specific ICD10 chapters. The LDA topic modeling analysis extracted 11 topics related to social determinants of health risk factors including financial status, abuse history, social support, risk of death, and mental health. In addition, the topic modeling approach captured the variation between different types of social work notes and across patients with different types of diseases or conditions. We demonstrated that social work notes contain rich, unique, and otherwise unobtainable information on an individual's SDoH.
Developing a general-purpose clinical language inference model from a large corpus of clinical notes
Oct 12, 2022
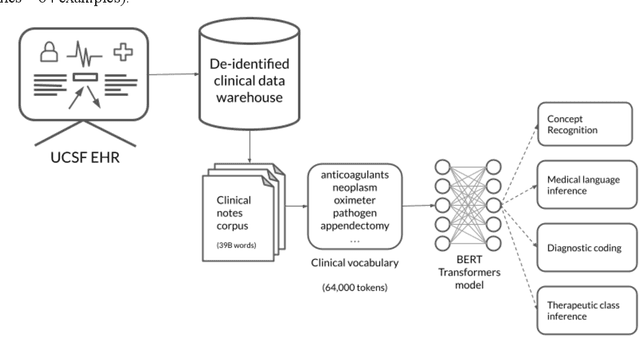
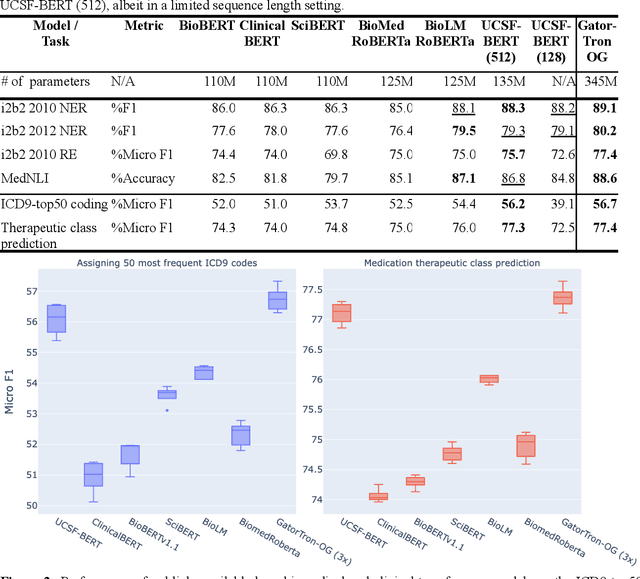
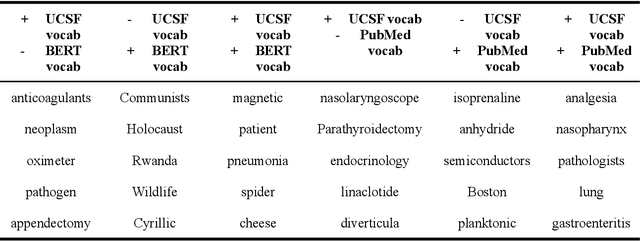
Abstract:Several biomedical language models have already been developed for clinical language inference. However, these models typically utilize general vocabularies and are trained on relatively small clinical corpora. We sought to evaluate the impact of using a domain-specific vocabulary and a large clinical training corpus on the performance of these language models in clinical language inference. We trained a Bidirectional Encoder Decoder from Transformers (BERT) model using a diverse, deidentified corpus of 75 million deidentified clinical notes authored at the University of California, San Francisco (UCSF). We evaluated this model on several clinical language inference benchmark tasks: clinical and temporal concept recognition, relation extraction and medical language inference. We also evaluated our model on two tasks using discharge summaries from UCSF: diagnostic code assignment and therapeutic class inference. Our model performs at par with the best publicly available biomedical language models of comparable sizes on the public benchmark tasks, and is significantly better than these models in a within-system evaluation on the two tasks using UCSF data. The use of in-domain vocabulary appears to improve the encoding of longer documents. The use of large clinical corpora appears to enhance document encoding and inferential accuracy. However, further research is needed to improve abbreviation resolution, and numerical, temporal, and implicitly causal inference.
Time Aggregation and Model Interpretation for Deep Multivariate Longitudinal Patient Outcome Forecasting Systems in Chronic Ambulatory Care
Nov 30, 2018
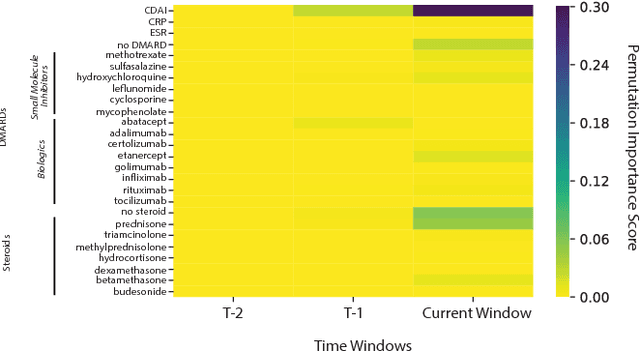
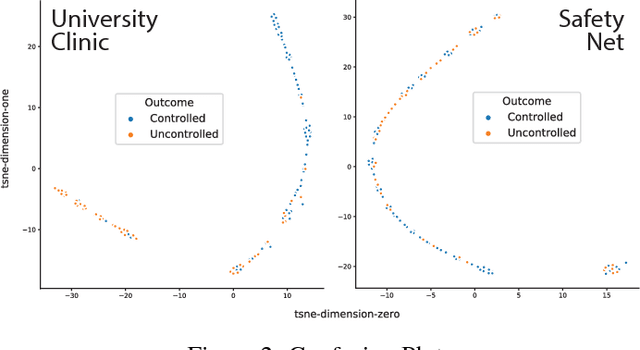
Abstract:Clinical data for ambulatory care, which accounts for 90% of the nations healthcare spending, is characterized by relatively small sample sizes of longitudinal data, unequal spacing between visits for each patient, with unequal numbers of data points collected across patients. While deep learning has become state-of-the-art for sequence modeling, it is unknown which methods of time aggregation may be best suited for these challenging temporal use cases. Additionally, deep models are often considered uninterpretable by physicians which may prevent the clinical adoption, even of well performing models. We show that time-distributed-dense layers combined with GRUs produce the most generalizable models. Furthermore, we provide a framework for the clinical interpretation of the models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge