Shenghuan Sun
Dr-LLaVA: Visual Instruction Tuning with Symbolic Clinical Grounding
May 29, 2024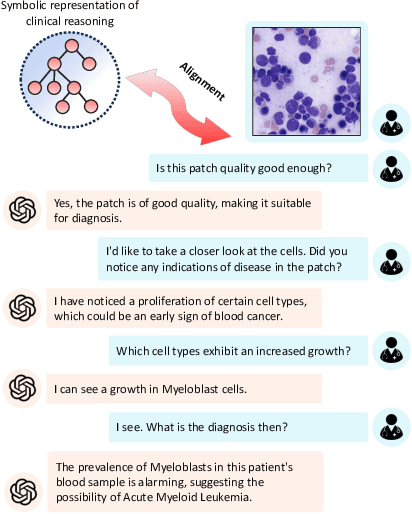
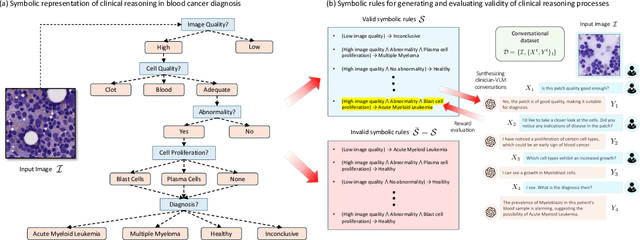
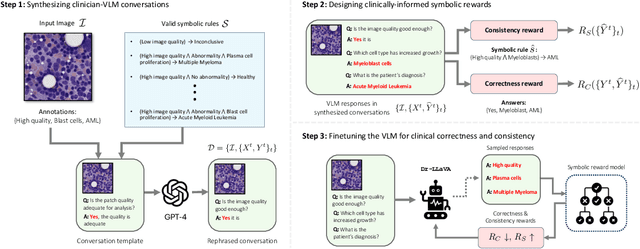
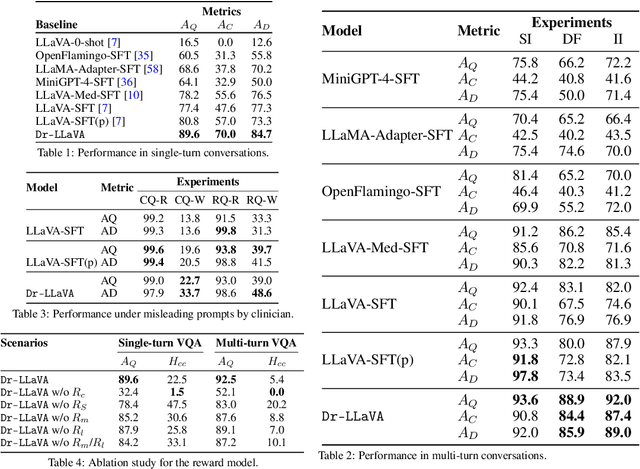
Abstract:Vision-Language Models (VLM) can support clinicians by analyzing medical images and engaging in natural language interactions to assist in diagnostic and treatment tasks. However, VLMs often exhibit "hallucinogenic" behavior, generating textual outputs not grounded in contextual multimodal information. This challenge is particularly pronounced in the medical domain, where we do not only require VLM outputs to be accurate in single interactions but also to be consistent with clinical reasoning and diagnostic pathways throughout multi-turn conversations. For this purpose, we propose a new alignment algorithm that uses symbolic representations of clinical reasoning to ground VLMs in medical knowledge. These representations are utilized to (i) generate GPT-4-guided visual instruction tuning data at scale, simulating clinician-VLM conversations with demonstrations of clinical reasoning, and (ii) create an automatic reward function that evaluates the clinical validity of VLM generations throughout clinician-VLM interactions. Our algorithm eliminates the need for human involvement in training data generation or reward model construction, reducing costs compared to standard reinforcement learning with human feedback (RLHF). We apply our alignment algorithm to develop Dr-LLaVA, a conversational VLM finetuned for analyzing bone marrow pathology slides, demonstrating strong performance in multi-turn medical conversations.
Aligning Synthetic Medical Images with Clinical Knowledge using Human Feedback
Jun 16, 2023



Abstract:Generative models capable of capturing nuanced clinical features in medical images hold great promise for facilitating clinical data sharing, enhancing rare disease datasets, and efficiently synthesizing annotated medical images at scale. Despite their potential, assessing the quality of synthetic medical images remains a challenge. While modern generative models can synthesize visually-realistic medical images, the clinical validity of these images may be called into question. Domain-agnostic scores, such as FID score, precision, and recall, cannot incorporate clinical knowledge and are, therefore, not suitable for assessing clinical sensibility. Additionally, there are numerous unpredictable ways in which generative models may fail to synthesize clinically plausible images, making it challenging to anticipate potential failures and manually design scores for their detection. To address these challenges, this paper introduces a pathologist-in-the-loop framework for generating clinically-plausible synthetic medical images. Starting with a diffusion model pretrained using real images, our framework comprises three steps: (1) evaluating the generated images by expert pathologists to assess whether they satisfy clinical desiderata, (2) training a reward model that predicts the pathologist feedback on new samples, and (3) incorporating expert knowledge into the diffusion model by using the reward model to inform a finetuning objective. We show that human feedback significantly improves the quality of synthetic images in terms of fidelity, diversity, utility in downstream applications, and plausibility as evaluated by experts.
Revealing the impact of social circumstances on the selection of cancer therapy through natural language processing of social work notes
Jun 16, 2023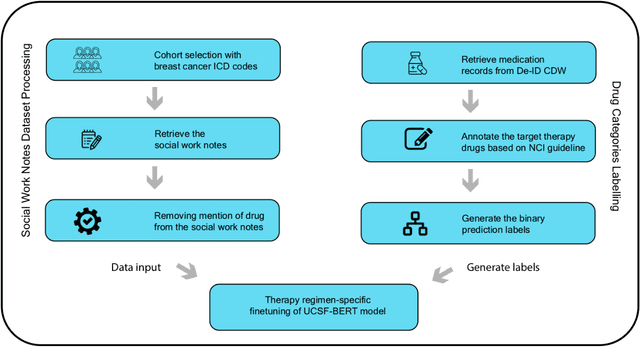
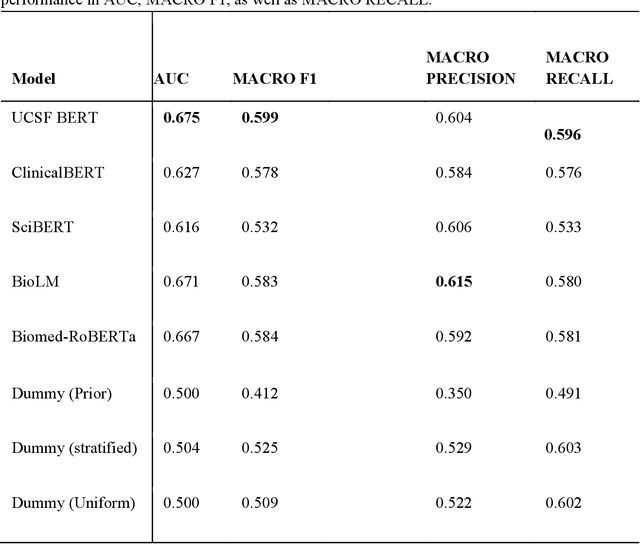
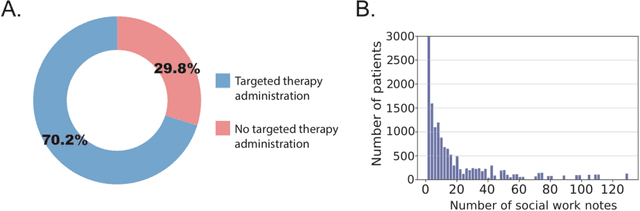
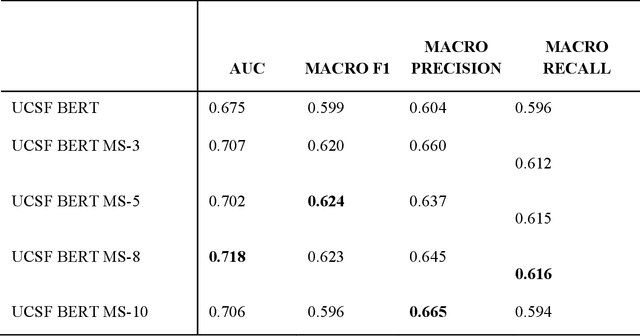
Abstract:We aimed to investigate the impact of social circumstances on cancer therapy selection using natural language processing to derive insights from social worker documentation. We developed and employed a Bidirectional Encoder Representations from Transformers (BERT) based approach, using a hierarchical multi-step BERT model (BERT-MS) to predict the prescription of targeted cancer therapy to patients based solely on documentation by clinical social workers. Our corpus included free-text clinical social work notes, combined with medication prescription information, for all patients treated for breast cancer. We conducted a feature importance analysis to pinpoint the specific social circumstances that impact cancer therapy selection. Using only social work notes, we consistently predicted the administration of targeted therapies, suggesting systematic differences in treatment selection exist due to non-clinical factors. The UCSF-BERT model, pretrained on clinical text at UCSF, outperformed other publicly available language models with an AUROC of 0.675 and a Macro F1 score of 0.599. The UCSF BERT-MS model, capable of leveraging multiple pieces of notes, surpassed the UCSF-BERT model in both AUROC and Macro-F1. Our feature importance analysis identified several clinically intuitive social determinants of health (SDOH) that potentially contribute to disparities in treatment. Our findings indicate that significant disparities exist among breast cancer patients receiving different types of therapies based on social determinants of health. Social work reports play a crucial role in understanding these disparities in clinical decision-making.
Topic Modeling on Clinical Social Work Notes for Exploring Social Determinants of Health Factors
Dec 02, 2022Abstract:Most research studying social determinants of health (SDoH) has focused on physician notes or structured elements of the electronic medical record (EMR). We hypothesize that clinical notes from social workers, whose role is to ameliorate social and economic factors, might provide a richer source of data on SDoH. We sought to perform topic modeling to identify robust topics of discussion within a large cohort of social work notes. We retrieved a diverse, deidentified corpus of 0.95 million clinical social work notes from 181,644 patients at the University of California, San Francisco. We used word frequency analysis and Latent Dirichlet Allocation (LDA) topic modeling analysis to characterize this corpus and identify potential topics of discussion. Word frequency analysis identified both medical and non-medical terms associated with specific ICD10 chapters. The LDA topic modeling analysis extracted 11 topics related to social determinants of health risk factors including financial status, abuse history, social support, risk of death, and mental health. In addition, the topic modeling approach captured the variation between different types of social work notes and across patients with different types of diseases or conditions. We demonstrated that social work notes contain rich, unique, and otherwise unobtainable information on an individual's SDoH.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge