Benjamin S. Glicksberg
Hasso Plattner Institute for Digital Health at Mount Sinai, Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, NY, USA
Cloud Platforms for Developing Generative AI Solutions: A Scoping Review of Tools and Services
Dec 08, 2024Abstract:Generative AI is transforming enterprise application development by enabling machines to create content, code, and designs. These models, however, demand substantial computational power and data management. Cloud computing addresses these needs by offering infrastructure to train, deploy, and scale generative AI models. This review examines cloud services for generative AI, focusing on key providers like Amazon Web Services (AWS), Microsoft Azure, Google Cloud, IBM Cloud, Oracle Cloud, and Alibaba Cloud. It compares their strengths, weaknesses, and impact on enterprise growth. We explore the role of high-performance computing (HPC), serverless architectures, edge computing, and storage in supporting generative AI. We also highlight the significance of data management, networking, and AI-specific tools in building and deploying these models. Additionally, the review addresses security concerns, including data privacy, compliance, and AI model protection. It assesses the performance and cost efficiency of various cloud providers and presents case studies from healthcare, finance, and entertainment. We conclude by discussing challenges and future directions, such as technical hurdles, vendor lock-in, sustainability, and regulatory issues. Put together, this work can serve as a guide for practitioners and researchers looking to adopt cloud-based generative AI solutions, serving as a valuable guide to navigating the intricacies of this evolving field.
Generation of a Compendium of Transcription Factor Cascades and Identification of Potential Therapeutic Targets using Graph Machine Learning
Nov 29, 2023



Abstract:Transcription factors (TFs) play a vital role in the regulation of gene expression thereby making them critical to many cellular processes. In this study, we used graph machine learning methods to create a compendium of TF cascades using data extracted from the STRING database. A TF cascade is a sequence of TFs that regulate each other, forming a directed path in the TF network. We constructed a knowledge graph of 81,488 unique TF cascades, with the longest cascade consisting of 62 TFs. Our results highlight the complex and intricate nature of TF interactions, where multiple TFs work together to regulate gene expression. We also identified 10 TFs with the highest regulatory influence based on centrality measurements, providing valuable information for researchers interested in studying specific TFs. Furthermore, our pathway enrichment analysis revealed significant enrichment of various pathways and functional categories, including those involved in cancer and other diseases, as well as those involved in development, differentiation, and cell signaling. The enriched pathways identified in this study may have potential as targets for therapeutic intervention in diseases associated with dysregulation of transcription factors. We have released the dataset, knowledge graph, and graphML methods for the TF cascades, and created a website to display the results, which can be accessed by researchers interested in using this dataset. Our study provides a valuable resource for understanding the complex network of interactions between TFs and their regulatory roles in cellular processes.
Beyond Low Earth Orbit: Biomonitoring, Artificial Intelligence, and Precision Space Health
Dec 22, 2021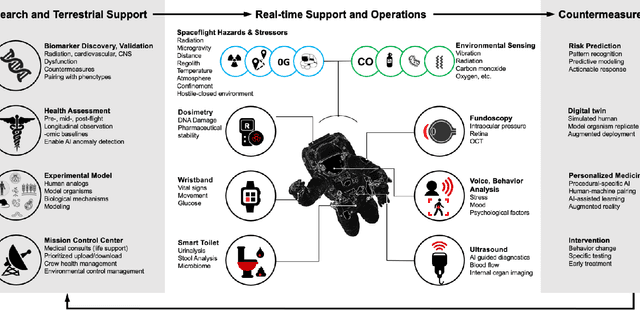
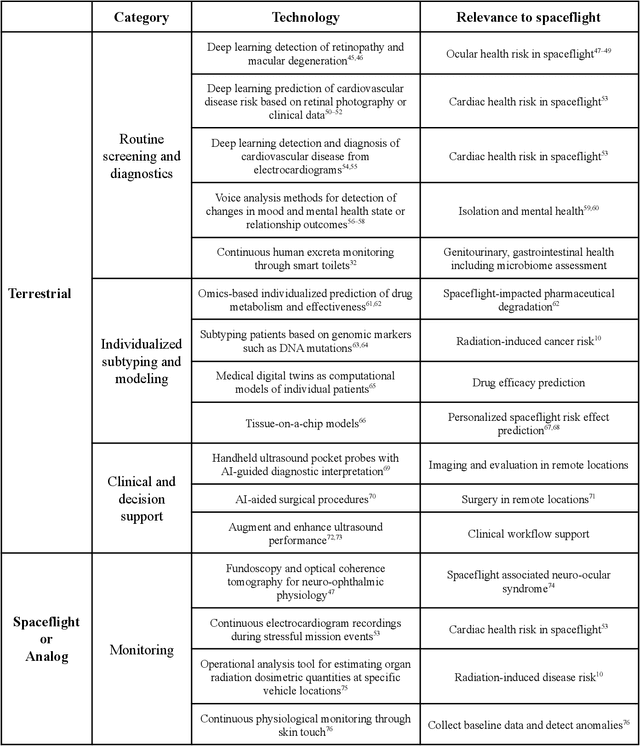
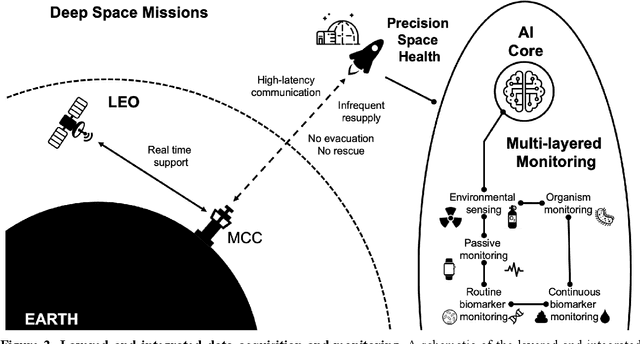
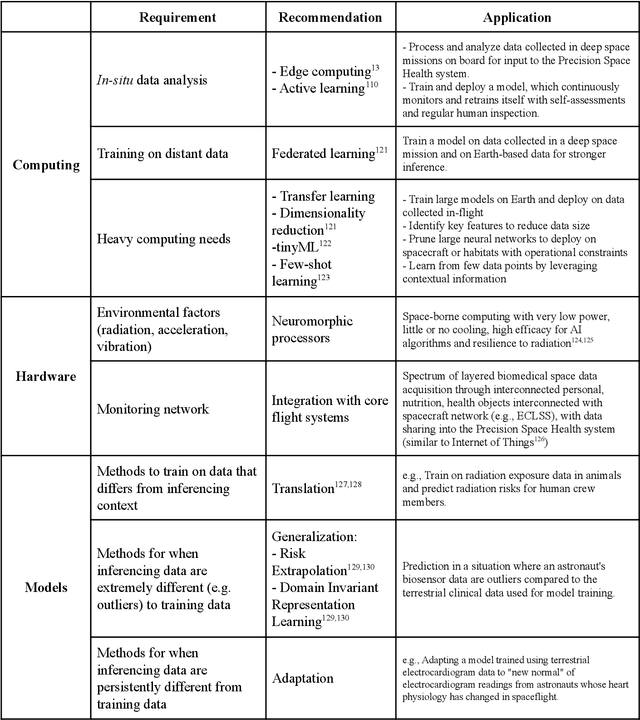
Abstract:Human space exploration beyond low Earth orbit will involve missions of significant distance and duration. To effectively mitigate myriad space health hazards, paradigm shifts in data and space health systems are necessary to enable Earth-independence, rather than Earth-reliance. Promising developments in the fields of artificial intelligence and machine learning for biology and health can address these needs. We propose an appropriately autonomous and intelligent Precision Space Health system that will monitor, aggregate, and assess biomedical statuses; analyze and predict personalized adverse health outcomes; adapt and respond to newly accumulated data; and provide preventive, actionable, and timely insights to individual deep space crew members and iterative decision support to their crew medical officer. Here we present a summary of recommendations from a workshop organized by the National Aeronautics and Space Administration, on future applications of artificial intelligence in space biology and health. In the next decade, biomonitoring technology, biomarker science, spacecraft hardware, intelligent software, and streamlined data management must mature and be woven together into a Precision Space Health system to enable humanity to thrive in deep space.
Beyond Low Earth Orbit: Biological Research, Artificial Intelligence, and Self-Driving Labs
Dec 22, 2021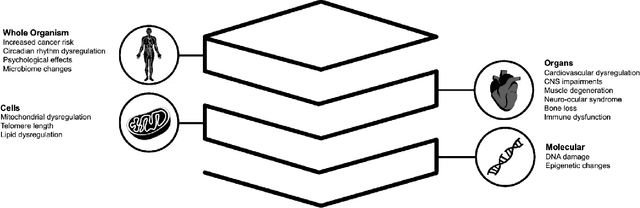
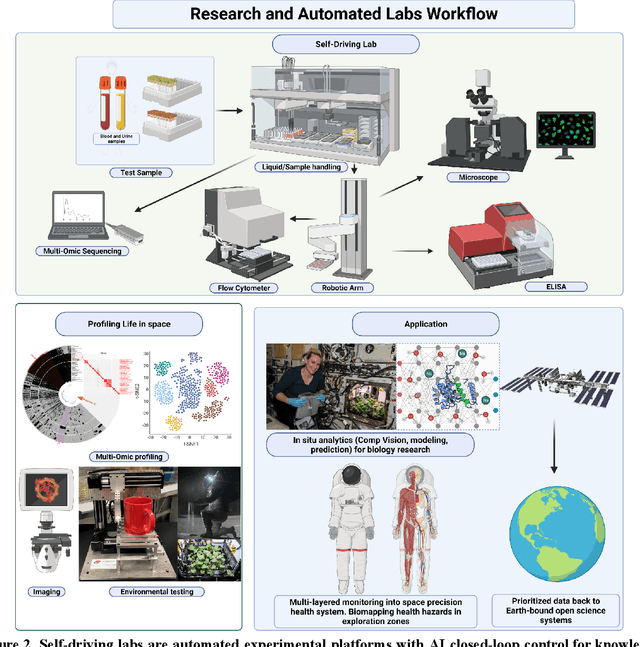
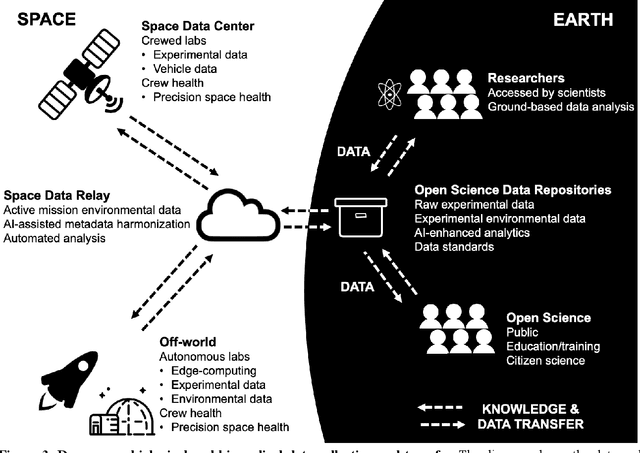
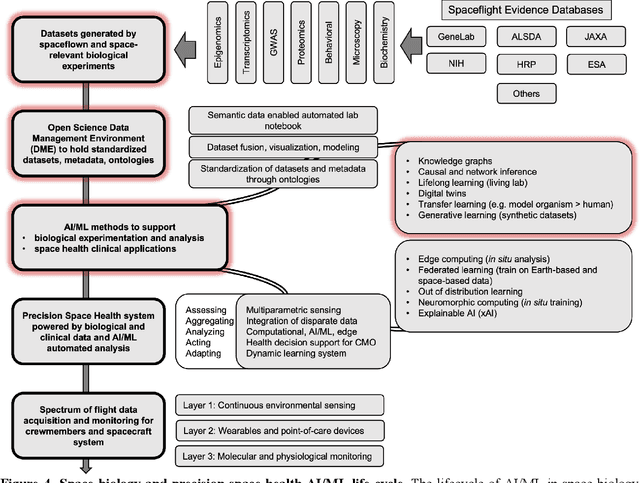
Abstract:Space biology research aims to understand fundamental effects of spaceflight on organisms, develop foundational knowledge to support deep space exploration, and ultimately bioengineer spacecraft and habitats to stabilize the ecosystem of plants, crops, microbes, animals, and humans for sustained multi-planetary life. To advance these aims, the field leverages experiments, platforms, data, and model organisms from both spaceborne and ground-analog studies. As research is extended beyond low Earth orbit, experiments and platforms must be maximally autonomous, light, agile, and intelligent to expedite knowledge discovery. Here we present a summary of recommendations from a workshop organized by the National Aeronautics and Space Administration on artificial intelligence, machine learning, and modeling applications which offer key solutions toward these space biology challenges. In the next decade, the synthesis of artificial intelligence into the field of space biology will deepen the biological understanding of spaceflight effects, facilitate predictive modeling and analytics, support maximally autonomous and reproducible experiments, and efficiently manage spaceborne data and metadata, all with the goal to enable life to thrive in deep space.
Contrastive Learning Improves Critical Event Prediction in COVID-19 Patients
Jan 11, 2021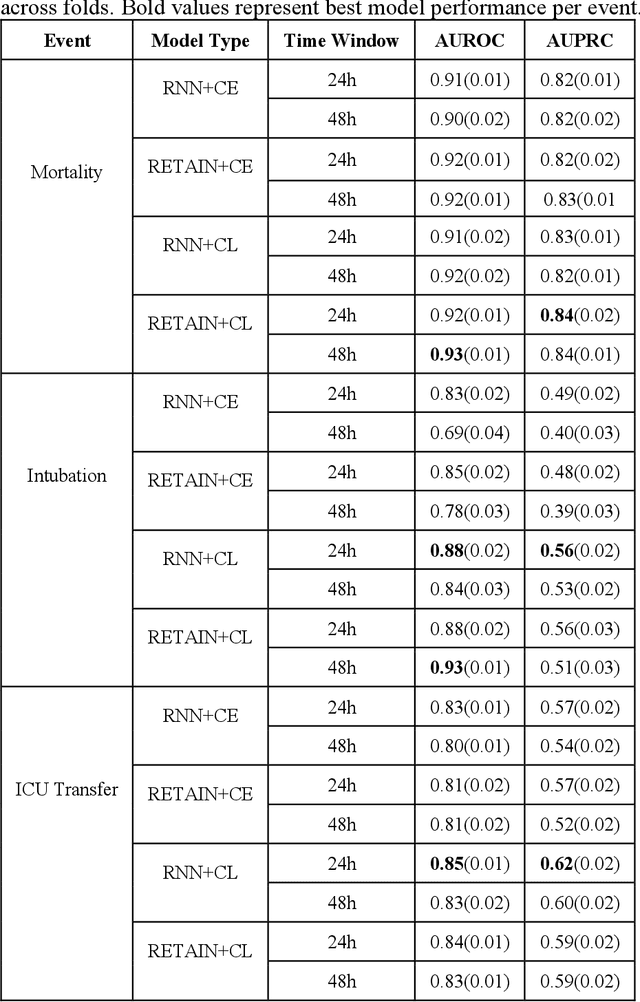
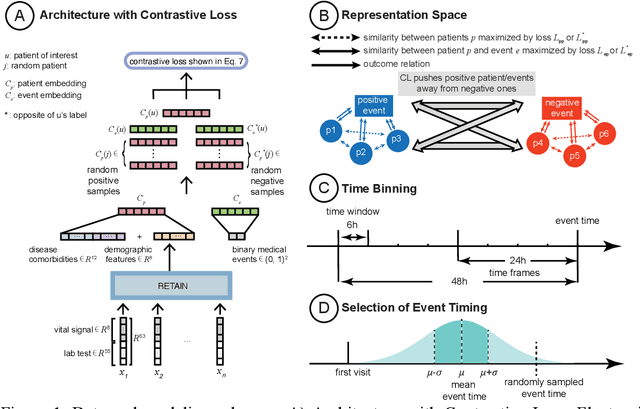
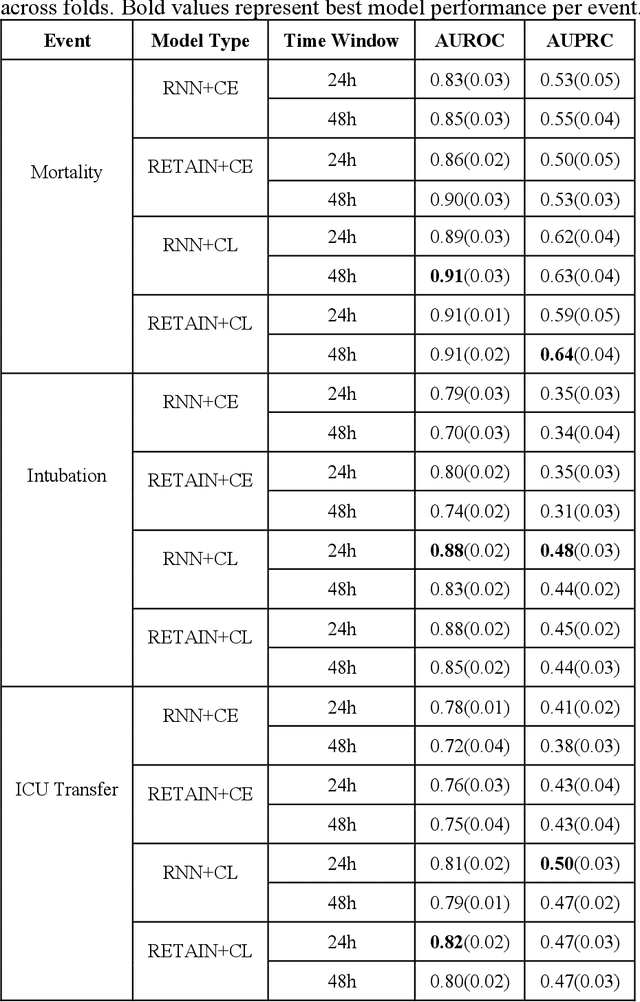
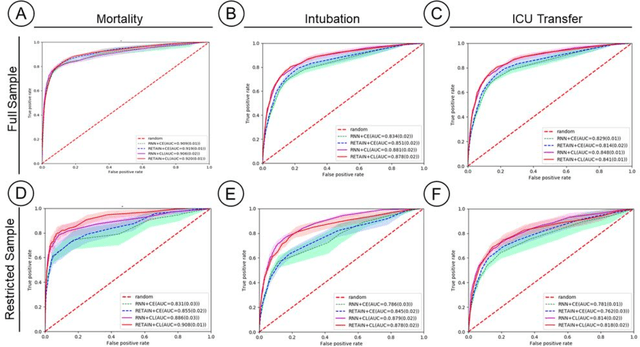
Abstract:Machine Learning (ML) models typically require large-scale, balanced training data to be robust, generalizable, and effective in the context of healthcare. This has been a major issue for developing ML models for the coronavirus-disease 2019 (COVID-19) pandemic where data is highly imbalanced, particularly within electronic health records (EHR) research. Conventional approaches in ML use cross-entropy loss (CEL) that often suffers from poor margin classification. For the first time, we show that contrastive loss (CL) improves the performance of CEL especially for imbalanced EHR data and the related COVID-19 analyses. This study has been approved by the Institutional Review Board at the Icahn School of Medicine at Mount Sinai. We use EHR data from five hospitals within the Mount Sinai Health System (MSHS) to predict mortality, intubation, and intensive care unit (ICU) transfer in hospitalized COVID-19 patients over 24 and 48 hour time windows. We train two sequential architectures (RNN and RETAIN) using two loss functions (CEL and CL). Models are tested on full sample data set which contain all available data and restricted data set to emulate higher class imbalance.CL models consistently outperform CEL models with the restricted data set on these tasks with differences ranging from 0.04 to 0.15 for AUPRC and 0.05 to 0.1 for AUROC. For the restricted sample, only the CL model maintains proper clustering and is able to identify important features, such as pulse oximetry. CL outperforms CEL in instances of severe class imbalance, on three EHR outcomes with respect to three performance metrics: predictive power, clustering, and feature importance. We believe that the developed CL framework can be expanded and used for EHR ML work in general.
Deep Learning with Heterogeneous Graph Embeddings for Mortality Prediction from Electronic Health Records
Dec 28, 2020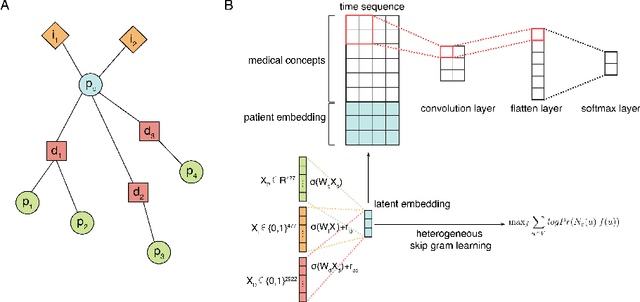
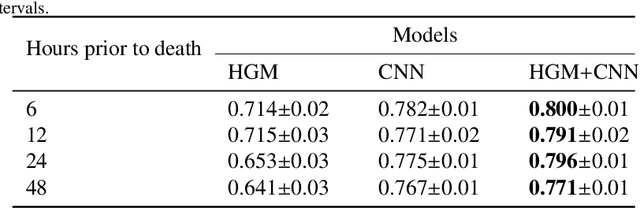
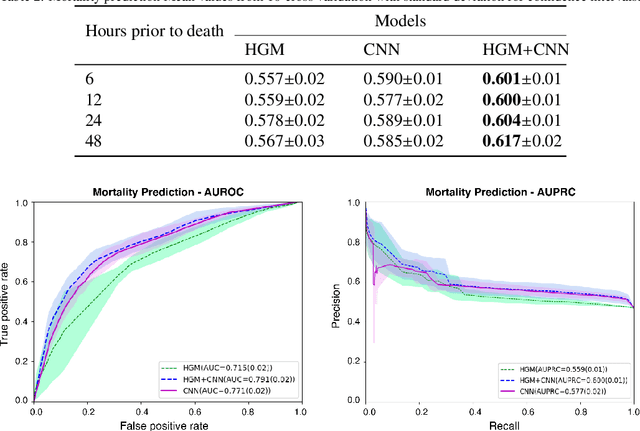
Abstract:Computational prediction of in-hospital mortality in the setting of an intensive care unit can help clinical practitioners to guide care and make early decisions for interventions. As clinical data are complex and varied in their structure and components, continued innovation of modeling strategies is required to identify architectures that can best model outcomes. In this work, we train a Heterogeneous Graph Model (HGM) on Electronic Health Record data and use the resulting embedding vector as additional information added to a Convolutional Neural Network (CNN) model for predicting in-hospital mortality. We show that the additional information provided by including time as a vector in the embedding captures the relationships between medical concepts, lab tests, and diagnoses, which enhances predictive performance. We find that adding HGM to a CNN model increases the mortality prediction accuracy up to 4\%. This framework serves as a foundation for future experiments involving different EHR data types on important healthcare prediction tasks.
Deep Representation Learning of Electronic Health Records to Unlock Patient Stratification at Scale
Mar 14, 2020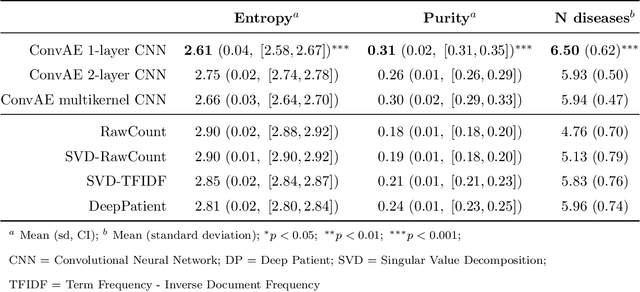
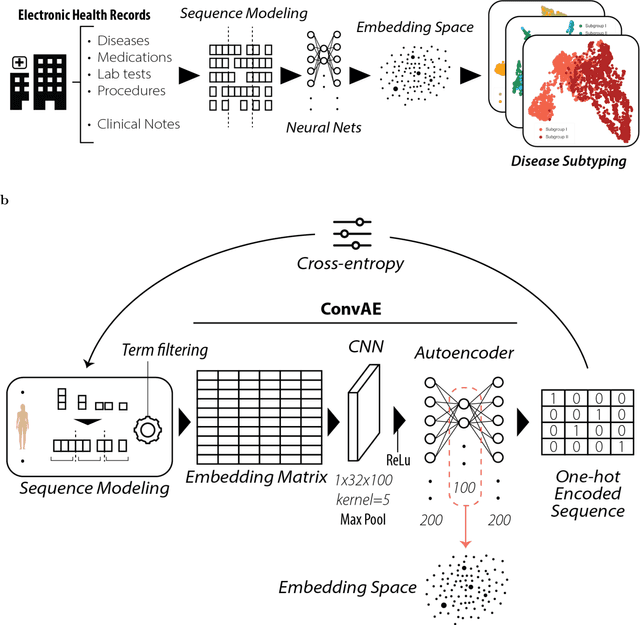
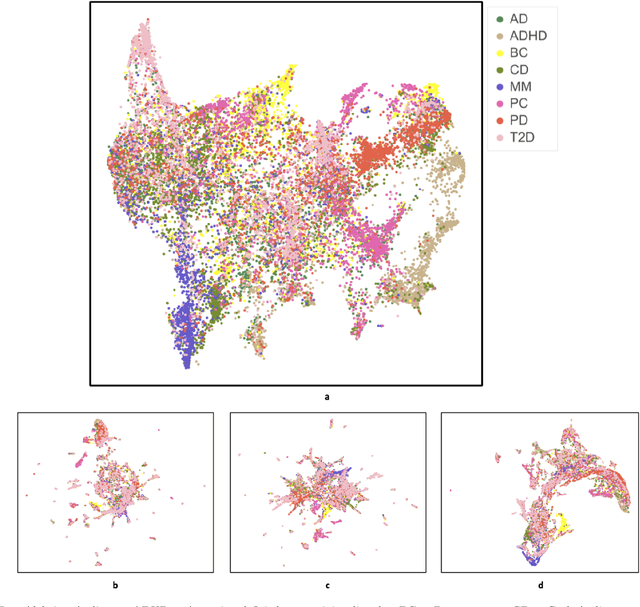
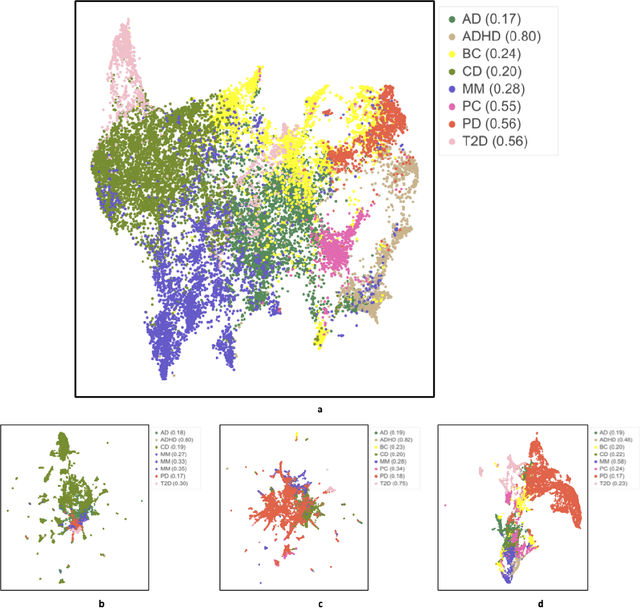
Abstract:Objective: Deriving disease subtypes from electronic health records (EHRs) can guide next-generation personalized medicine. However, challenges in summarizing and representing patient data prevent widespread practice of scalable EHR-based stratification analysis. Here, we present a novel unsupervised framework based on deep learning to process heterogeneous EHRs and derive patient representations that can efficiently and effectively enable patient stratification at scale. Materials and methods: We considered EHRs of $1,608,741$ patients from a diverse hospital cohort comprising of a total of $57,464$ clinical concepts. We introduce a representation learning model based on word embeddings, convolutional neural networks and autoencoders (i.e., "ConvAE") to transform patient trajectories into low-dimensional latent vectors. We evaluated these representations as broadly enabling patient stratification by applying hierarchical clustering to different multi-disease and disease-specific patient cohorts. Results: ConvAE significantly outperformed several common baselines in a clustering task to identify patients with different complex conditions, with $2.61$ entropy and $0.31$ purity average scores. When applied to stratify patients within a certain condition, ConvAE led to various clinically relevant subtypes for different disorders, including type 2 diabetes, Parkinson's disease and Alzheimer's disease, largely related to comorbidities, disease progression, and symptom severity. Conclusions: Patient representations derived from modeling EHRs with ConvAE can help develop personalized medicine therapeutic strategies and better understand varying etiologies in heterogeneous sub-populations.
Time Aggregation and Model Interpretation for Deep Multivariate Longitudinal Patient Outcome Forecasting Systems in Chronic Ambulatory Care
Nov 30, 2018
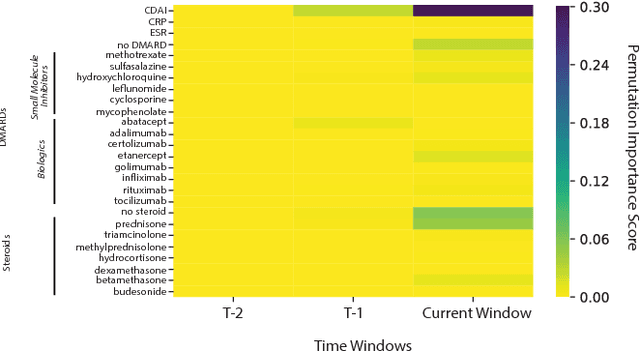
Abstract:Clinical data for ambulatory care, which accounts for 90% of the nations healthcare spending, is characterized by relatively small sample sizes of longitudinal data, unequal spacing between visits for each patient, with unequal numbers of data points collected across patients. While deep learning has become state-of-the-art for sequence modeling, it is unknown which methods of time aggregation may be best suited for these challenging temporal use cases. Additionally, deep models are often considered uninterpretable by physicians which may prevent the clinical adoption, even of well performing models. We show that time-distributed-dense layers combined with GRUs produce the most generalizable models. Furthermore, we provide a framework for the clinical interpretation of the models.
Deep Learning Predicts Hip Fracture using Confounding Patient and Healthcare Variables
Nov 08, 2018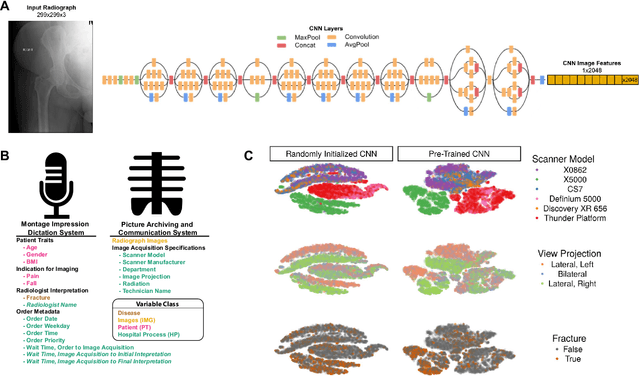
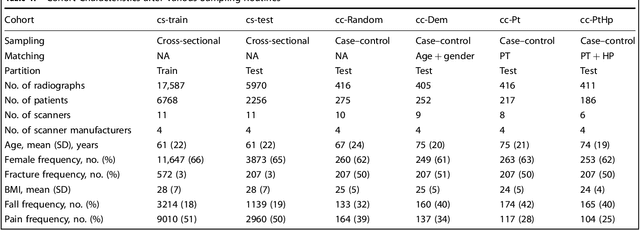
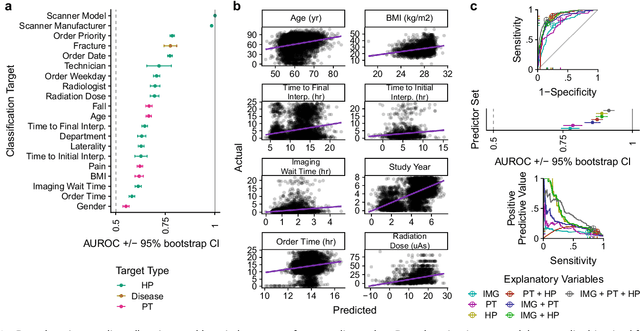
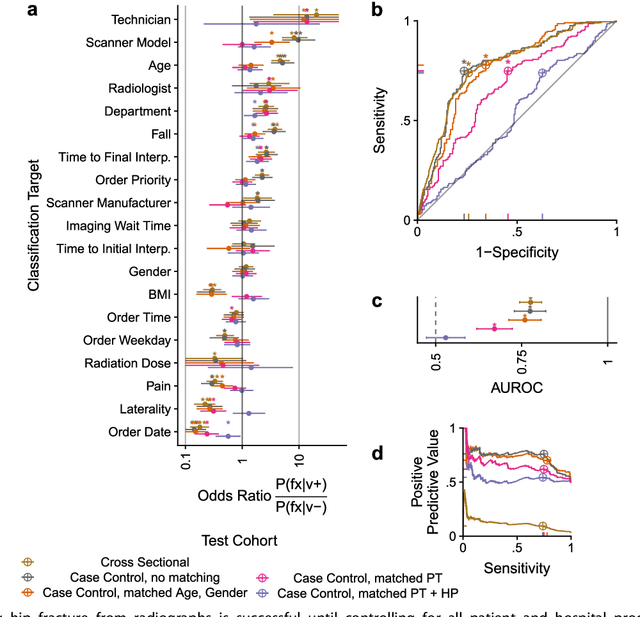
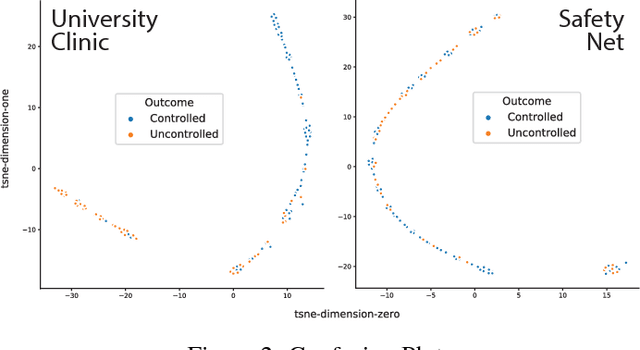
Abstract:Hip fractures are a leading cause of death and disability among older adults. Hip fractures are also the most commonly missed diagnosis on pelvic radiographs. Computer-Aided Diagnosis (CAD) algorithms have shown promise for helping radiologists detect fractures, but the image features underpinning their predictions are notoriously difficult to understand. In this study, we trained deep learning models on 17,587 radiographs to classify fracture, five patient traits, and 14 hospital process variables. All 20 variables could be predicted from a radiograph (p < 0.05), with the best performances on scanner model (AUC=1.00), scanner brand (AUC=0.98), and whether the order was marked "priority" (AUC=0.79). Fracture was predicted moderately well from the image (AUC=0.78) and better when combining image features with patient data (AUC=0.86, p=2e-9) or patient data plus hospital process features (AUC=0.91, p=1e-21). The model performance on a test set with matched patient variables was significantly lower than a random test set (AUC=0.67, p=0.003); and when the test set was matched on patient and image acquisition variables, the model performed randomly (AUC=0.52, 95% CI 0.46-0.58), indicating that these variables were the main source of the model's predictive ability overall. We also used Naive Bayes to combine evidence from image models with patient and hospital data and found their inclusion improved performance, but that this approach was nevertheless inferior to directly modeling all variables. If CAD algorithms are inexplicably leveraging patient and process variables in their predictions, it is unclear how radiologists should interpret their predictions in the context of other known patient data. Further research is needed to illuminate deep learning decision processes so that computers and clinicians can effectively cooperate.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge