Ankit Bhardwaj
Self-Regulating Cars: Automating Traffic Control in Free Flow Road Networks
Jun 13, 2025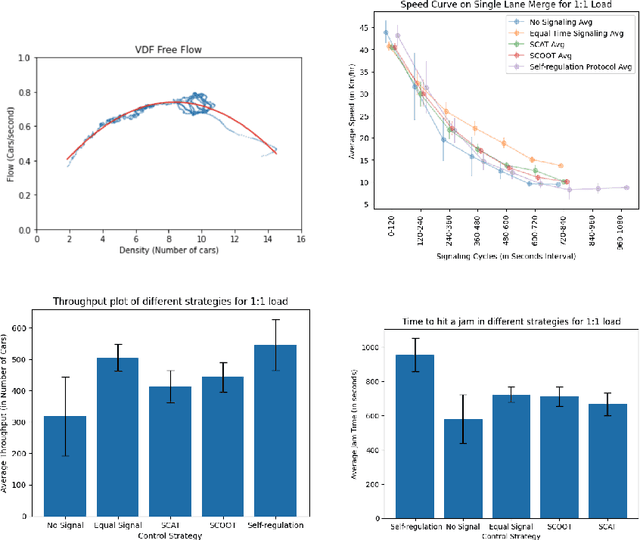
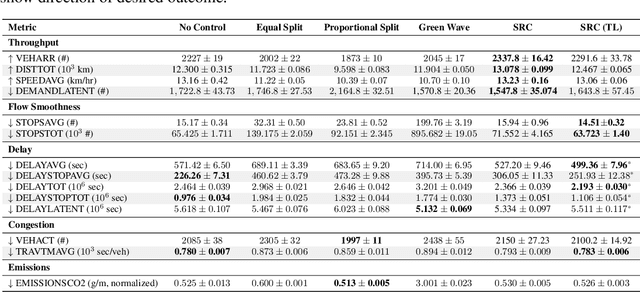
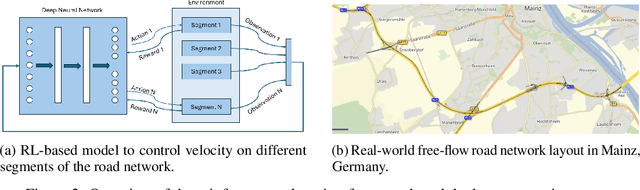
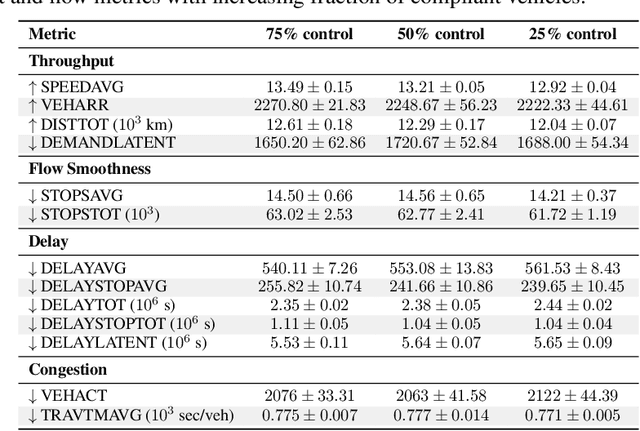
Abstract:Free-flow road networks, such as suburban highways, are increasingly experiencing traffic congestion due to growing commuter inflow and limited infrastructure. Traditional control mechanisms, such as traffic signals or local heuristics, are ineffective or infeasible in these high-speed, signal-free environments. We introduce self-regulating cars, a reinforcement learning-based traffic control protocol that dynamically modulates vehicle speeds to optimize throughput and prevent congestion, without requiring new physical infrastructure. Our approach integrates classical traffic flow theory, gap acceptance models, and microscopic simulation into a physics-informed RL framework. By abstracting roads into super-segments, the agent captures emergent flow dynamics and learns robust speed modulation policies from instantaneous traffic observations. Evaluated in the high-fidelity PTV Vissim simulator on a real-world highway network, our method improves total throughput by 5%, reduces average delay by 13%, and decreases total stops by 3% compared to the no-control setting. It also achieves smoother, congestion-resistant flow while generalizing across varied traffic patterns, demonstrating its potential for scalable, ML-driven traffic management.
Discovering Hidden Pollution Hotspots Using Sparse Sensor Measurements
Oct 05, 2024Abstract:Effective air pollution management in urban areas relies on both monitoring and mitigation strategies, yet high costs often limit sensor networks to a few key pollution hotspots. In this paper, we show that New Delhi's public sensor network is insufficient for identifying all pollution hotspots. To address this, we augmented the city's network with 28 low-cost sensors, monitoring PM 2.5 concentrations over 30 months (May 2018 to November 2020). Our analysis uncovered 189 additional hotspots, supplementing the 660 already detected by the government network. We observed that Space-Time Kriging with limited but accurate sensor data provides a more robust and generalizable approach for identifying these hotspots, as compared to deep learning models that require large amounts of fine-grained multi-modal data (emissions inventory, meteorology, etc.) which was not reliably, frequently and accurately available in the New Delhi context. Using Space-Time Kriging, we achieved 98% precision and 95.4% recall in detecting hotspots with 50% sensor failure. Furthermore, this method proved effective in predicting hotspots in areas without sensors, achieving 95.3% precision and 88.5% recall in the case of 50% missing sensors. Our findings revealed that a significant portion of New Delhi's population, around 23 million people, was exposed to pollution hotspots for at least half of the study period. We also identified areas beyond the reach of the public sensor network that should be prioritized for pollution control. These results highlight the need for more comprehensive monitoring networks and suggest Space-Time Kriging as a viable solution for cities facing similar resource constraints.
Packrat: Automatic Reconfiguration for Latency Minimization in CPU-based DNN Serving
Nov 30, 2023Abstract:In this paper, we investigate how to push the performance limits of serving Deep Neural Network (DNN) models on CPU-based servers. Specifically, we observe that while intra-operator parallelism across multiple threads is an effective way to reduce inference latency, it provides diminishing returns. Our primary insight is that instead of running a single instance of a model with all available threads on a server, running multiple instances each with smaller batch sizes and fewer threads for intra-op parallelism can provide lower inference latency. However, the right configuration is hard to determine manually since it is workload- (DNN model and batch size used by the serving system) and deployment-dependent (number of CPU cores on server). We present Packrat, a new serving system for online inference that given a model and batch size ($B$) algorithmically picks the optimal number of instances ($i$), the number of threads each should be allocated ($t$), and the batch sizes each should operate on ($b$) that minimizes latency. Packrat is built as an extension to TorchServe and supports online reconfigurations to avoid serving downtime. Averaged across a range of batch sizes, Packrat improves inference latency by 1.43$\times$ to 1.83$\times$ on a range of commonly used DNNs.
Generation of a Compendium of Transcription Factor Cascades and Identification of Potential Therapeutic Targets using Graph Machine Learning
Nov 29, 2023



Abstract:Transcription factors (TFs) play a vital role in the regulation of gene expression thereby making them critical to many cellular processes. In this study, we used graph machine learning methods to create a compendium of TF cascades using data extracted from the STRING database. A TF cascade is a sequence of TFs that regulate each other, forming a directed path in the TF network. We constructed a knowledge graph of 81,488 unique TF cascades, with the longest cascade consisting of 62 TFs. Our results highlight the complex and intricate nature of TF interactions, where multiple TFs work together to regulate gene expression. We also identified 10 TFs with the highest regulatory influence based on centrality measurements, providing valuable information for researchers interested in studying specific TFs. Furthermore, our pathway enrichment analysis revealed significant enrichment of various pathways and functional categories, including those involved in cancer and other diseases, as well as those involved in development, differentiation, and cell signaling. The enriched pathways identified in this study may have potential as targets for therapeutic intervention in diseases associated with dysregulation of transcription factors. We have released the dataset, knowledge graph, and graphML methods for the TF cascades, and created a website to display the results, which can be accessed by researchers interested in using this dataset. Our study provides a valuable resource for understanding the complex network of interactions between TFs and their regulatory roles in cellular processes.
Weakly-Supervised Deep Learning Model for Prostate Cancer Diagnosis and Gleason Grading of Histopathology Images
Dec 25, 2022Abstract:Prostate cancer is the most common cancer in men worldwide and the second leading cause of cancer death in the United States. One of the prognostic features in prostate cancer is the Gleason grading of histopathology images. The Gleason grade is assigned based on tumor architecture on Hematoxylin and Eosin (H&E) stained whole slide images (WSI) by the pathologists. This process is time-consuming and has known interobserver variability. In the past few years, deep learning algorithms have been used to analyze histopathology images, delivering promising results for grading prostate cancer. However, most of the algorithms rely on the fully annotated datasets which are expensive to generate. In this work, we proposed a novel weakly-supervised algorithm to classify prostate cancer grades. The proposed algorithm consists of three steps: (1) extracting discriminative areas in a histopathology image by employing the Multiple Instance Learning (MIL) algorithm based on Transformers, (2) representing the image by constructing a graph using the discriminative patches, and (3) classifying the image into its Gleason grades by developing a Graph Convolutional Neural Network (GCN) based on the gated attention mechanism. We evaluated our algorithm using publicly available datasets, including TCGAPRAD, PANDA, and Gleason 2019 challenge datasets. We also cross validated the algorithm on an independent dataset. Results show that the proposed model achieved state-of-the-art performance in the Gleason grading task in terms of accuracy, F1 score, and cohen-kappa. The code is available at https://github.com/NabaviLab/Prostate-Cancer.
Predicting Treatment Adherence of Tuberculosis Patients at Scale
Nov 15, 2022



Abstract:Tuberculosis (TB), an infectious bacterial disease, is a significant cause of death, especially in low-income countries, with an estimated ten million new cases reported globally in $2020$. While TB is treatable, non-adherence to the medication regimen is a significant cause of morbidity and mortality. Thus, proactively identifying patients at risk of dropping off their medication regimen enables corrective measures to mitigate adverse outcomes. Using a proxy measure of extreme non-adherence and a dataset of nearly $700,000$ patients from four states in India, we formulate and solve the machine learning (ML) problem of early prediction of non-adherence based on a custom rank-based metric. We train ML models and evaluate against baselines, achieving a $\sim 100\%$ lift over rule-based baselines and $\sim 214\%$ over a random classifier, taking into account country-wide large-scale future deployment. We deal with various issues in the process, including data quality, high-cardinality categorical data, low target prevalence, distribution shift, variation across cohorts, algorithmic fairness, and the need for robustness and explainability. Our findings indicate that risk stratification of non-adherent patients is a viable, deployable-at-scale ML solution. As the official AI partner of India's Central TB Division, we are working on multiple city and state-level pilots with the goal of pan-India deployment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge