Ariful Azad
Shapley-Value-Based Graph Sparsification for GNN Inference
Jul 28, 2025Abstract:Graph sparsification is a key technique for improving inference efficiency in Graph Neural Networks by removing edges with minimal impact on predictions. GNN explainability methods generate local importance scores, which can be aggregated into global scores for graph sparsification. However, many explainability methods produce only non-negative scores, limiting their applicability for sparsification. In contrast, Shapley value based methods assign both positive and negative contributions to node predictions, offering a theoretically robust and fair allocation of importance by evaluating many subsets of graphs. Unlike gradient-based or perturbation-based explainers, Shapley values enable better pruning strategies that preserve influential edges while removing misleading or adversarial connections. Our approach shows that Shapley value-based graph sparsification maintains predictive performance while significantly reducing graph complexity, enhancing both interpretability and efficiency in GNN inference.
PLANETALIGN: A Comprehensive Python Library for Benchmarking Network Alignment
May 27, 2025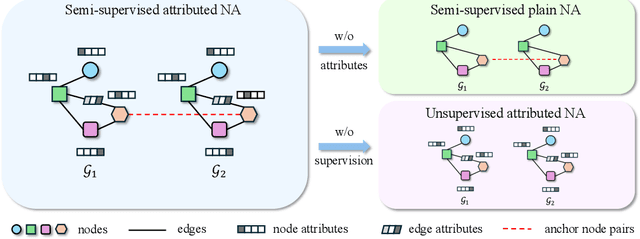
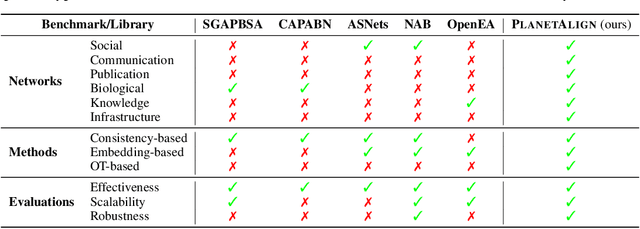
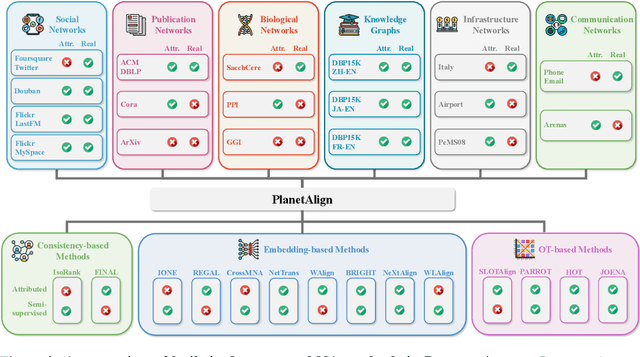
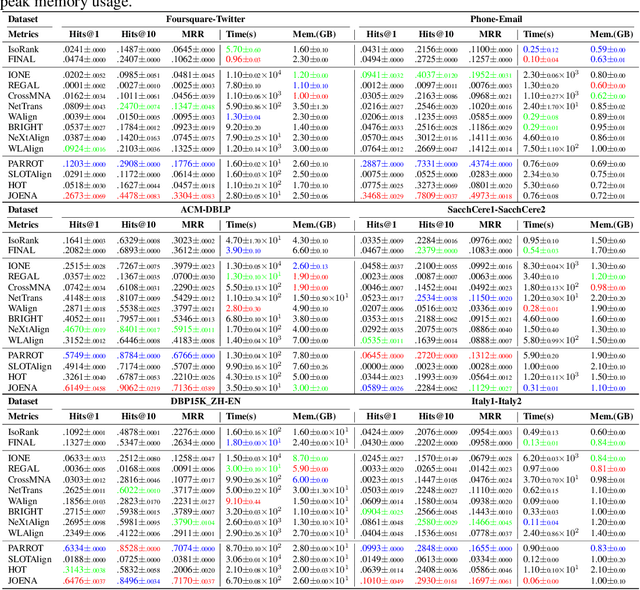
Abstract:Network alignment (NA) aims to identify node correspondence across different networks and serves as a critical cornerstone behind various downstream multi-network learning tasks. Despite growing research in NA, there lacks a comprehensive library that facilitates the systematic development and benchmarking of NA methods. In this work, we introduce PLANETALIGN, a comprehensive Python library for network alignment that features a rich collection of built-in datasets, methods, and evaluation pipelines with easy-to-use APIs. Specifically, PLANETALIGN integrates 18 datasets and 14 NA methods with extensible APIs for easy use and development of NA methods. Our standardized evaluation pipeline encompasses a wide range of metrics, enabling a systematic assessment of the effectiveness, scalability, and robustness of NA methods. Through extensive comparative studies, we reveal practical insights into the strengths and limitations of existing NA methods. We hope that PLANETALIGN can foster a deeper understanding of the NA problem and facilitate the development and benchmarking of more effective, scalable, and robust methods in the future. The source code of PLANETALIGN is available at https://github.com/yq-leo/PlanetAlign.
SparseTransX: Efficient Training of Translation-Based Knowledge Graph Embeddings Using Sparse Matrix Operations
Feb 26, 2025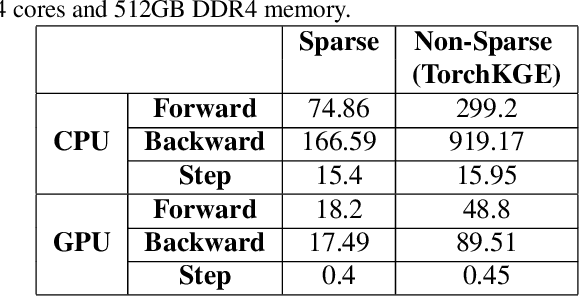
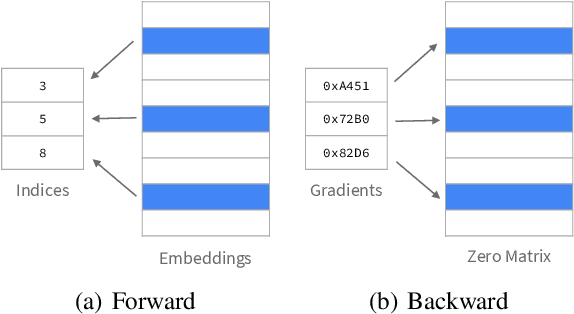
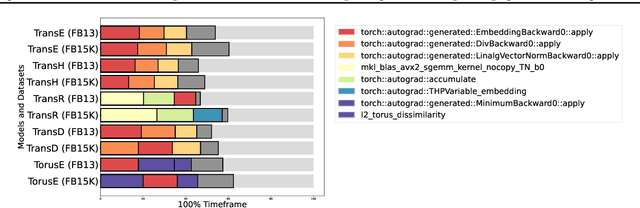
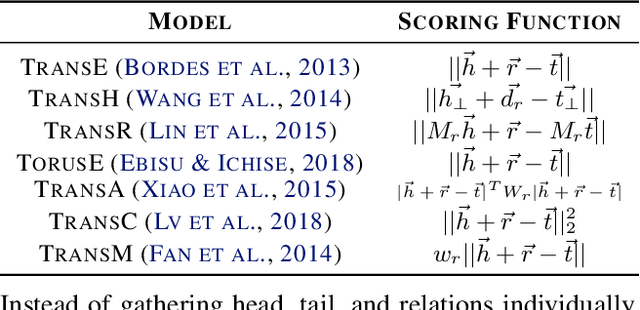
Abstract:Knowledge graph (KG) learning offers a powerful framework for generating new knowledge and making inferences. Training KG embedding can take a significantly long time, especially for larger datasets. Our analysis shows that the gradient computation of embedding is one of the dominant functions in the translation-based KG embedding training loop. We address this issue by replacing the core embedding computation with SpMM (Sparse-Dense Matrix Multiplication) kernels. This allows us to unify multiple scatter (and gather) operations as a single operation, reducing training time and memory usage. We create a general framework for training KG models using sparse kernels and implement four models, namely TransE, TransR, TransH, and TorusE. Our sparse implementations exhibit up to 5.3x speedup on the CPU and up to 4.2x speedup on the GPU with a significantly low GPU memory footprint. The speedups are consistent across large and small datasets for a given model. Our proposed sparse approach can be extended to accelerate other translation-based (such as TransC, TransM, etc.) and non-translational (such as DistMult, ComplEx, RotatE, etc.) models as well.
Publication Trends in Artificial Intelligence Conferences: The Rise of Super Prolific Authors
Nov 28, 2024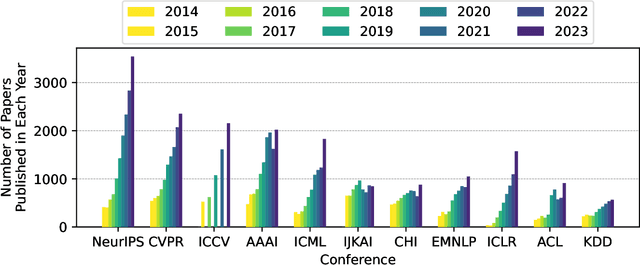
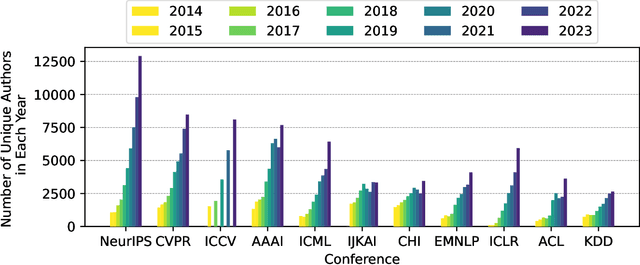
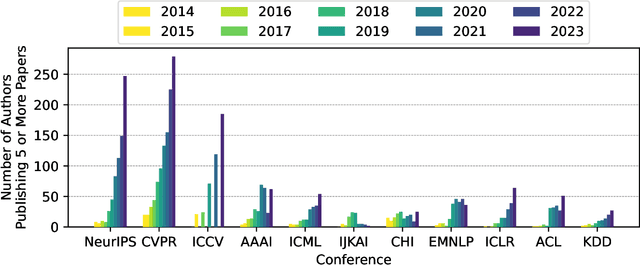
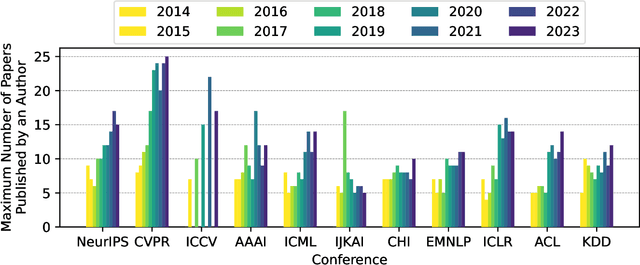
Abstract:Papers published in top conferences contribute influential discoveries that are reshaping the landscape of modern Artificial Intelligence (AI). We analyzed 87,137 papers from 11 AI conferences to examine publication trends over the past decade. Our findings reveal a consistent increase in both the number of papers and authors, reflecting the growing interest in AI research. We also observed a rise in prolific researchers who publish dozens of papers at the same conference each year. In light of this analysis, the AI research community should consider revisiting authorship policies, addressing equity concerns, and evaluating the workload of junior researchers to foster a more sustainable and inclusive research environment.
Parallel Algorithms for Median Consensus Clustering in Complex Networks
Aug 21, 2024Abstract:We develop an algorithm that finds the consensus of many different clustering solutions of a graph. We formulate the problem as a median set partitioning problem and propose a greedy optimization technique. Unlike other approaches that find median set partitions, our algorithm takes graph structure into account and finds a comparable quality solution much faster than the other approaches. For graphs with known communities, our consensus partition captures the actual community structure more accurately than alternative approaches. To make it applicable to large graphs, we remove sequential dependencies from our algorithm and design a parallel algorithm. Our parallel algorithm achieves 35x speedup when utilizing 64 processing cores for large real-world graphs from single-cell experiments.
iSpLib: A Library for Accelerating Graph Neural Networks using Auto-tuned Sparse Operations
Mar 21, 2024Abstract:Core computations in Graph Neural Network (GNN) training and inference are often mapped to sparse matrix operations such as sparse-dense matrix multiplication (SpMM). These sparse operations are harder to optimize by manual tuning because their performance depends significantly on the sparsity of input graphs, GNN models, and computing platforms. To address this challenge, we present iSpLib, a PyTorch-based C++ library equipped with auto-tuned sparse operations. iSpLib expedites GNN training with a cache-enabled backpropagation that stores intermediate matrices in local caches. The library offers a user-friendly Python plug-in that allows users to take advantage of our optimized PyTorch operations out-of-the-box for any existing linear algebra-based PyTorch implementation of popular GNNs (Graph Convolution Network, GraphSAGE, Graph Inference Network, etc.) with only two lines of additional code. We demonstrate that iSpLib obtains up to 27x overall training speedup compared to the equivalent PyTorch 2.1.0 and PyTorch Geometric 2.4.0 implementations on the CPU. Our library is publicly available at https://github.com/HipGraph/iSpLib (https://doi.org/10.5281/zenodo.10806511).
GNNShap: Fast and Accurate GNN Explanations using Shapley Values
Jan 09, 2024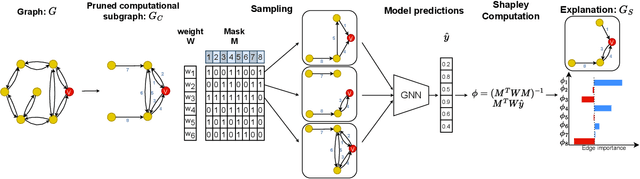
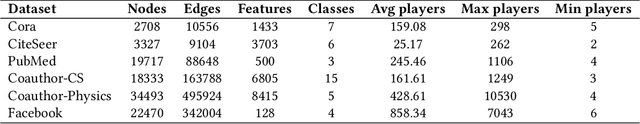
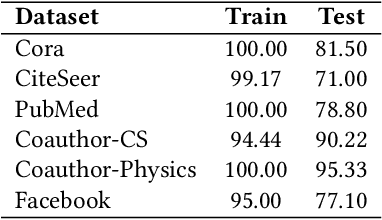
Abstract:Graph neural networks (GNNs) are popular machine learning models for graphs with many applications across scientific domains. However, GNNs are considered black box models, and it is challenging to understand how the model makes predictions. Game theory-based Shapley value approaches are popular explanation methods in other domains but are not well-studied for graphs. Some studies have proposed Shapley value-based GNN explanations, yet they have several limitations: they consider limited samples to approximate Shapley values; some mainly focus on small and large coalition sizes, and they are an order of magnitude slower than other explanation methods, making them inapplicable to even moderate-size graphs. In this work, we propose GNNShap, which provides explanations for edges since they provide more natural explanations for graphs and more fine-grained explanations. We overcome the limitations by sampling from all coalition sizes, parallelizing the sampling on GPUs, and speeding up model predictions by batching. GNNShap gives better fidelity scores and faster explanations than baselines on real-world datasets.
Triple Sparsification of Graph Convolutional Networks without Sacrificing the Accuracy
Aug 06, 2022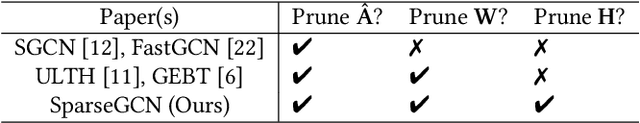
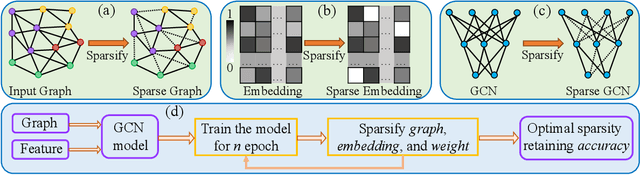
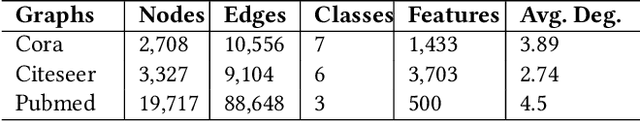
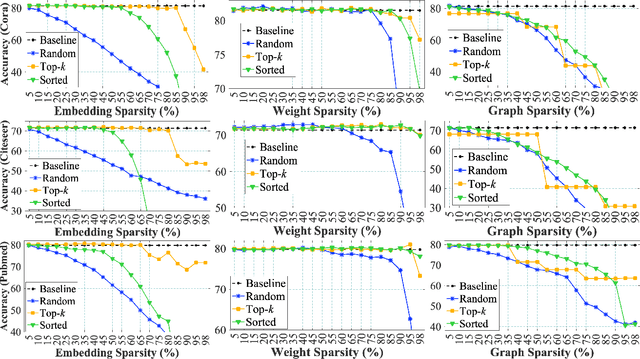
Abstract:Graph Neural Networks (GNNs) are widely used to perform different machine learning tasks on graphs. As the size of the graphs grows, and the GNNs get deeper, training and inference time become costly in addition to the memory requirement. Thus, without sacrificing accuracy, graph sparsification, or model compression becomes a viable approach for graph learning tasks. A few existing techniques only study the sparsification of graphs and GNN models. In this paper, we develop a SparseGCN pipeline to study all possible sparsification in GNN. We provide a theoretical analysis and empirically show that it can add up to 11.6\% additional sparsity to the embedding matrix without sacrificing the accuracy of the commonly used benchmark graph datasets.
Graphical Games for UAV Swarm Control Under Time-Varying Communication Networks
May 04, 2022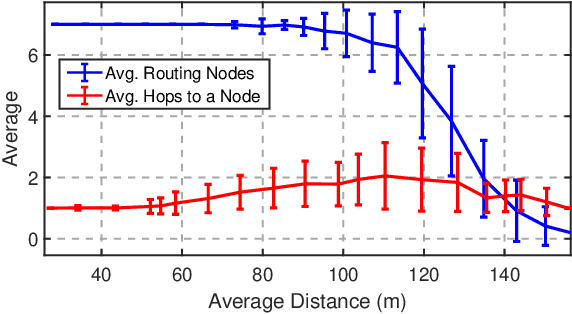
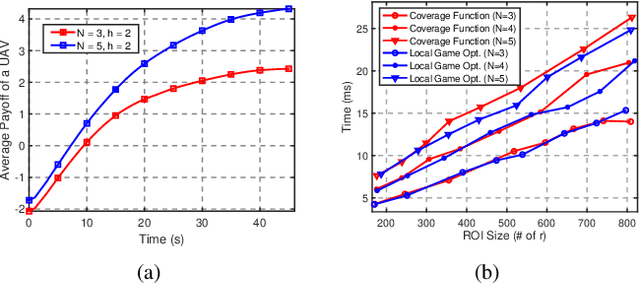
Abstract:We propose a unified framework for coordinating Unmanned Aerial Vehicle (UAV) swarms operating under time-varying communication networks. Our framework builds on the concept of graphical games, which we argue provides a compelling paradigm to subsume the interaction structures found in networked UAV swarms thanks to the shared local neighborhood properties. We present a general-sum, factorizable payoff function for cooperative UAV swarms based on the aggregated local states and yield a Nash equilibrium for the stage games. Further, we propose a decomposition-based approach to solve stage-graphical games in a scalable and decentralized fashion by approximating virtual, mean neighborhoods. Finally, we discuss extending the proposed framework toward general-sum stochastic games by leveraging deep Q-learning and model-predictive control.
MarkovGNN: Graph Neural Networks on Markov Diffusion
Feb 05, 2022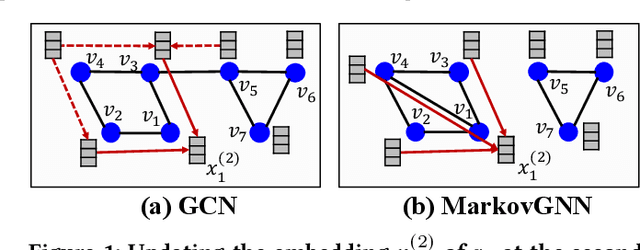
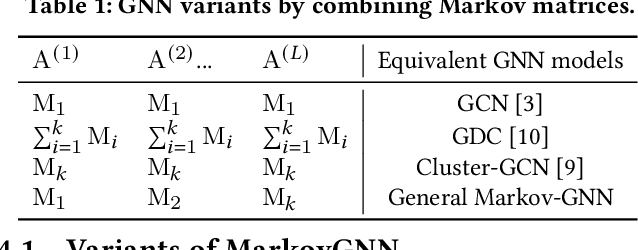
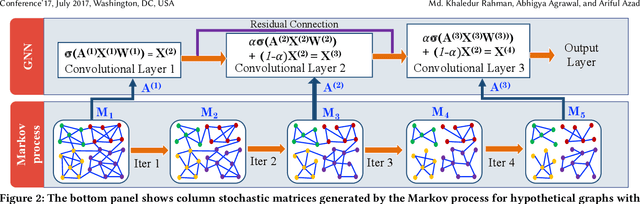
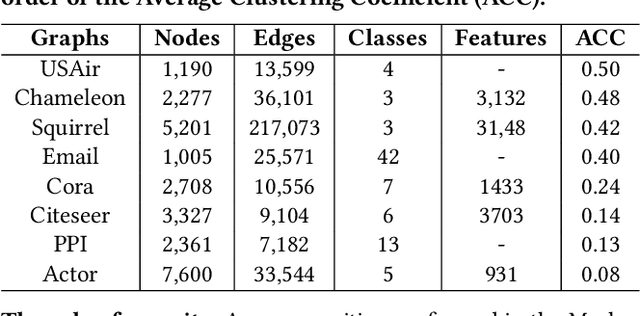
Abstract:Most real-world networks contain well-defined community structures where nodes are densely connected internally within communities. To learn from these networks, we develop MarkovGNN that captures the formation and evolution of communities directly in different convolutional layers. Unlike most Graph Neural Networks (GNNs) that consider a static graph at every layer, MarkovGNN generates different stochastic matrices using a Markov process and then uses these community-capturing matrices in different layers. MarkovGNN is a general approach that could be used with most existing GNNs. We experimentally show that MarkovGNN outperforms other GNNs for clustering, node classification, and visualization tasks. The source code of MarkovGNN is publicly available at \url{https://github.com/HipGraph/MarkovGNN}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge