Isotta Landi
Bruno Kessler Foundation, Trento, Italy, University of Trento, Italy
The Point of View of a Sentiment: Towards Clinician Bias Detection in Psychiatric Notes
May 31, 2024



Abstract:In psychiatry, negative patient descriptions and stigmatizing language can contribute to healthcare disparities in two ways: (1) read by patients they can harm their trust and engagement with the medical center; (2) read by future providers they may negatively influence the future perspective of a patient. By leveraging large language models, this work aims to identify the sentiment expressed in psychiatric clinical notes based on the reader's point of view. Extracting sentences from the Mount Sinai Health System's large and diverse clinical notes, we used prompts and in-context learning to adapt three large language models (GPT-3.5, Llama 2, Mistral) to classify the sentiment conveyed by the sentences according to the provider or non-provider point of view. Results showed that GPT-3.5 aligns best to provider point of view, whereas Mistral aligns best to non-provider point of view.
Extracting Social Support and Social Isolation Information from Clinical Psychiatry Notes: Comparing a Rule-based NLP System and a Large Language Model
Mar 25, 2024Abstract:Background: Social support (SS) and social isolation (SI) are social determinants of health (SDOH) associated with psychiatric outcomes. In electronic health records (EHRs), individual-level SS/SI is typically documented as narrative clinical notes rather than structured coded data. Natural language processing (NLP) algorithms can automate the otherwise labor-intensive process of data extraction. Data and Methods: Psychiatric encounter notes from Mount Sinai Health System (MSHS, n=300) and Weill Cornell Medicine (WCM, n=225) were annotated and established a gold standard corpus. A rule-based system (RBS) involving lexicons and a large language model (LLM) using FLAN-T5-XL were developed to identify mentions of SS and SI and their subcategories (e.g., social network, instrumental support, and loneliness). Results: For extracting SS/SI, the RBS obtained higher macro-averaged f-scores than the LLM at both MSHS (0.89 vs. 0.65) and WCM (0.85 vs. 0.82). For extracting subcategories, the RBS also outperformed the LLM at both MSHS (0.90 vs. 0.62) and WCM (0.82 vs. 0.81). Discussion and Conclusion: Unexpectedly, the RBS outperformed the LLMs across all metrics. Intensive review demonstrates that this finding is due to the divergent approach taken by the RBS and LLM. The RBS were designed and refined to follow the same specific rules as the gold standard annotations. Conversely, the LLM were more inclusive with categorization and conformed to common English-language understanding. Both approaches offer advantages and are made available open-source for future testing.
Generative Large Language Models are autonomous practitioners of evidence-based medicine
Jan 05, 2024



Abstract:Background: Evidence-based medicine (EBM) is fundamental to modern clinical practice, requiring clinicians to continually update their knowledge and apply the best clinical evidence in patient care. The practice of EBM faces challenges due to rapid advancements in medical research, leading to information overload for clinicians. The integration of artificial intelligence (AI), specifically Generative Large Language Models (LLMs), offers a promising solution towards managing this complexity. Methods: This study involved the curation of real-world clinical cases across various specialties, converting them into .json files for analysis. LLMs, including proprietary models like ChatGPT 3.5 and 4, Gemini Pro, and open-source models like LLaMA v2 and Mixtral-8x7B, were employed. These models were equipped with tools to retrieve information from case files and make clinical decisions similar to how clinicians must operate in the real world. Model performance was evaluated based on correctness of final answer, judicious use of tools, conformity to guidelines, and resistance to hallucinations. Results: GPT-4 was most capable of autonomous operation in a clinical setting, being generally more effective in ordering relevant investigations and conforming to clinical guidelines. Limitations were observed in terms of model ability to handle complex guidelines and diagnostic nuances. Retrieval Augmented Generation made recommendations more tailored to patients and healthcare systems. Conclusions: LLMs can be made to function as autonomous practitioners of evidence-based medicine. Their ability to utilize tooling can be harnessed to interact with the infrastructure of a real-world healthcare system and perform the tasks of patient management in a guideline directed manner. Prompt engineering may help to further enhance this potential and transform healthcare for the clinician and the patient.
Keyword-optimized Template Insertion for Clinical Information Extraction via Prompt-based Learning
Oct 31, 2023Abstract:Clinical note classification is a common clinical NLP task. However, annotated data-sets are scarse. Prompt-based learning has recently emerged as an effective method to adapt pre-trained models for text classification using only few training examples. A critical component of prompt design is the definition of the template (i.e. prompt text). The effect of template position, however, has been insufficiently investigated. This seems particularly important in the clinical setting, where task-relevant information is usually sparse in clinical notes. In this study we develop a keyword-optimized template insertion method (KOTI) and show how optimizing position can improve performance on several clinical tasks in a zero-shot and few-shot training setting.
reval: a Python package to determine the best number of clusters with stability-based relative clustering validation
Aug 27, 2020



Abstract:Determining the number of clusters that best partitions a dataset can be a challenging task because of 1) the lack of a priori information within an unsupervised learning framework; and 2) the absence of a unique clustering validation approach to evaluate clustering solutions. Here we present reval: a Python package that leverages stability-based relative clustering validation methods to determine best clustering solutions. Statistical software, both in R and Python, usually rely on internal validation metrics, such as the silhouette index, to select the number of clusters that best fits the data. Meanwhile, open-source software solutions that easily implement relative clustering techniques are lacking. Internal validation methods exploit characteristics of the data itself to produce a result, whereas relative approaches attempt to leverage the unknown underlying distribution of data points looking for a replicable and generalizable clustering solution. The implementation of relative validation solutions can further the theory of clustering by enriching the already available methods that can be used to investigate clustering results in different situations and for different data distributions. This work aims at contributing to this effort by developing a stability-based method that selects the best clustering solution as the one that replicates, via supervised learning, on unseen subsets of data. The package works with multiple clustering and classification algorithms, hence allowing further assessment of the stability of different clustering mechanisms.
Deep Representation Learning of Electronic Health Records to Unlock Patient Stratification at Scale
Mar 14, 2020
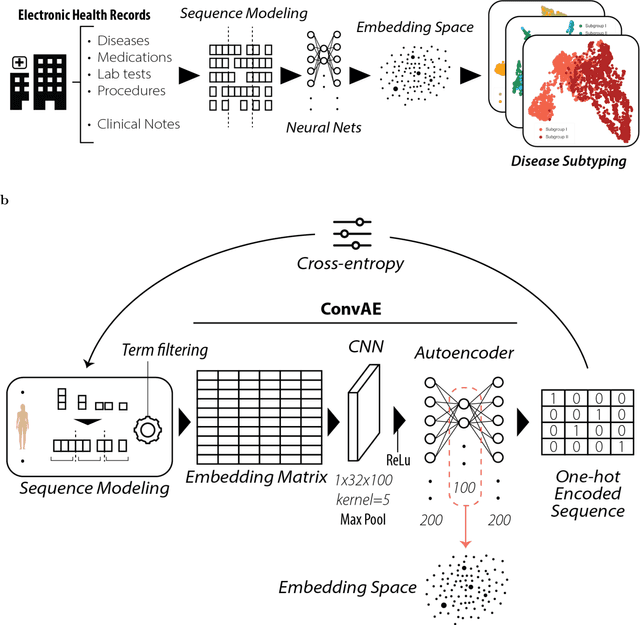
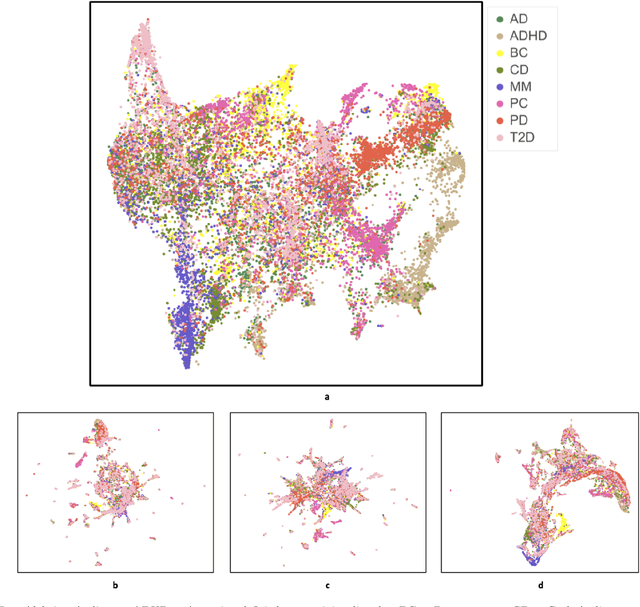
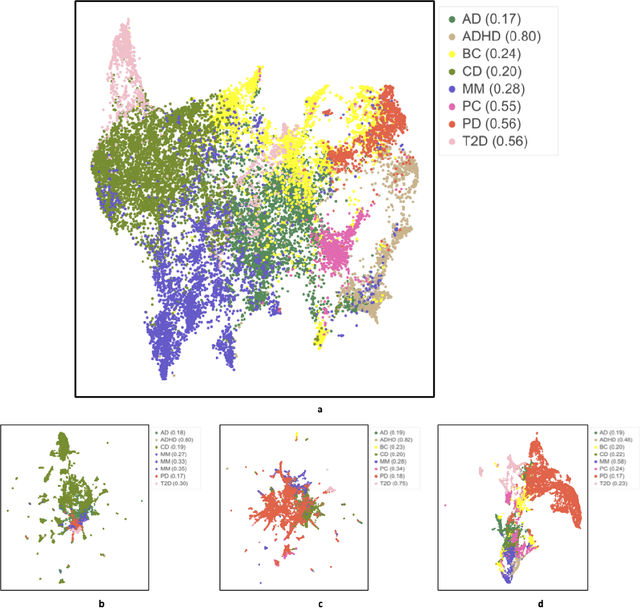
Abstract:Objective: Deriving disease subtypes from electronic health records (EHRs) can guide next-generation personalized medicine. However, challenges in summarizing and representing patient data prevent widespread practice of scalable EHR-based stratification analysis. Here, we present a novel unsupervised framework based on deep learning to process heterogeneous EHRs and derive patient representations that can efficiently and effectively enable patient stratification at scale. Materials and methods: We considered EHRs of $1,608,741$ patients from a diverse hospital cohort comprising of a total of $57,464$ clinical concepts. We introduce a representation learning model based on word embeddings, convolutional neural networks and autoencoders (i.e., "ConvAE") to transform patient trajectories into low-dimensional latent vectors. We evaluated these representations as broadly enabling patient stratification by applying hierarchical clustering to different multi-disease and disease-specific patient cohorts. Results: ConvAE significantly outperformed several common baselines in a clustering task to identify patients with different complex conditions, with $2.61$ entropy and $0.31$ purity average scores. When applied to stratify patients within a certain condition, ConvAE led to various clinically relevant subtypes for different disorders, including type 2 diabetes, Parkinson's disease and Alzheimer's disease, largely related to comorbidities, disease progression, and symptom severity. Conclusions: Patient representations derived from modeling EHRs with ConvAE can help develop personalized medicine therapeutic strategies and better understand varying etiologies in heterogeneous sub-populations.
Convolutional neural networks for structured omics: OmicsCNN and the OmicsConv layer
Oct 16, 2017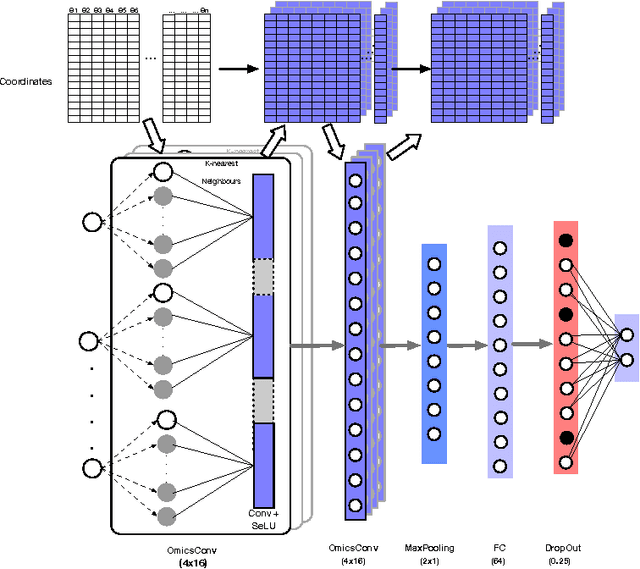

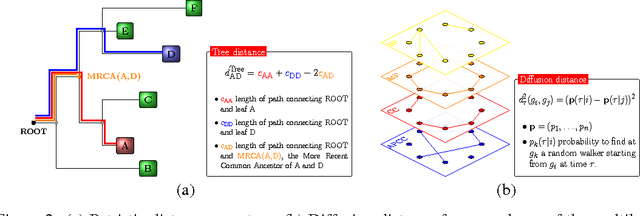
Abstract:Convolutional Neural Networks (CNNs) are a popular deep learning architecture widely applied in different domains, in particular in classifying over images, for which the concept of convolution with a filter comes naturally. Unfortunately, the requirement of a distance (or, at least, of a neighbourhood function) in the input feature space has so far prevented its direct use on data types such as omics data. However, a number of omics data are metrizable, i.e., they can be endowed with a metric structure, enabling to adopt a convolutional based deep learning framework, e.g., for prediction. We propose a generalized solution for CNNs on omics data, implemented through a dedicated Keras layer. In particular, for metagenomics data, a metric can be derived from the patristic distance on the phylogenetic tree. For transcriptomics data, we combine Gene Ontology semantic similarity and gene co-expression to define a distance; the function is defined through a multilayer network where 3 layers are defined by the GO mutual semantic similarity while the fourth one by gene co-expression. As a general tool, feature distance on omics data is enabled by OmicsConv, a novel Keras layer, obtaining OmicsCNN, a dedicated deep learning framework. Here we demonstrate OmicsCNN on gut microbiota sequencing data, for Inflammatory Bowel Disease (IBD) 16S data, first on synthetic data and then a metagenomics collection of gut microbiota of 222 IBD patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge