Diego Fioravanti
Tangles: From Weak to Strong Clustering
Jun 25, 2020



Abstract:We introduce a new approach to clustering by using tangles, a tool that originates in mathematical graph theory. Given a collection of "weak partitions" of a data set, tangles provide a framework to aggregate these weak partitions such that they "point in the direction of a cluster". As a result, a cluster is softly characterized by a set of consistent pointers. This mechanism provides a highly flexible way of solving soft clustering problems in a variety of setups, ranging from questionnaires over community detection in graphs to clustering points in metric spaces. Conceptually, tangles have many intriguing properties: (1) Similar to boosting, which combines many weak classifiers to a strong classifier, tangles provide a formal way to combine many weak partitions to obtain few strong clusters. (2) In terms of computational complexity, tangles allow us to use simple, fast algorithms to produce the weak partitions. The complexity of identifying the strong partitions is dominated by the number of weak partitions, not the number of data points, leading to an interesting trade-off between the two. (3) If the weak partitions are interpretable, so are the strong partitions induced by the tangles, resulting in one of the rare algorithms to produce interpretable clusters. (4) The output of tangles is of a hierarchical nature, inducing the notion of a soft dendrogram that can be helpful in data visualization.
Convolutional neural networks for structured omics: OmicsCNN and the OmicsConv layer
Oct 16, 2017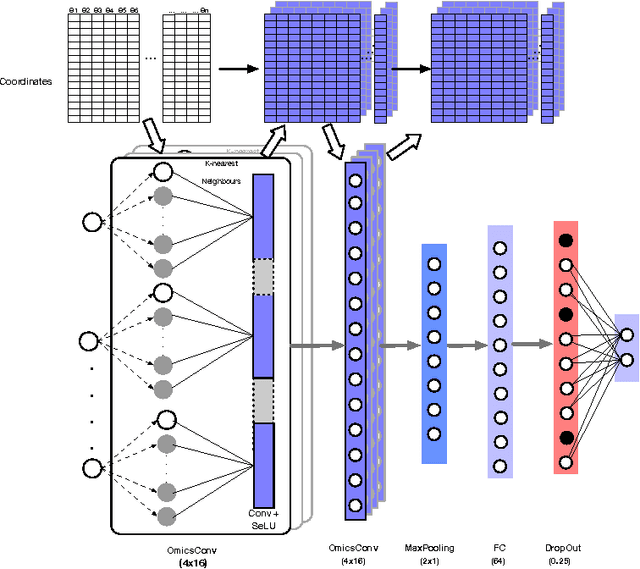

Abstract:Convolutional Neural Networks (CNNs) are a popular deep learning architecture widely applied in different domains, in particular in classifying over images, for which the concept of convolution with a filter comes naturally. Unfortunately, the requirement of a distance (or, at least, of a neighbourhood function) in the input feature space has so far prevented its direct use on data types such as omics data. However, a number of omics data are metrizable, i.e., they can be endowed with a metric structure, enabling to adopt a convolutional based deep learning framework, e.g., for prediction. We propose a generalized solution for CNNs on omics data, implemented through a dedicated Keras layer. In particular, for metagenomics data, a metric can be derived from the patristic distance on the phylogenetic tree. For transcriptomics data, we combine Gene Ontology semantic similarity and gene co-expression to define a distance; the function is defined through a multilayer network where 3 layers are defined by the GO mutual semantic similarity while the fourth one by gene co-expression. As a general tool, feature distance on omics data is enabled by OmicsConv, a novel Keras layer, obtaining OmicsCNN, a dedicated deep learning framework. Here we demonstrate OmicsCNN on gut microbiota sequencing data, for Inflammatory Bowel Disease (IBD) 16S data, first on synthetic data and then a metagenomics collection of gut microbiota of 222 IBD patients.
Phylogenetic Convolutional Neural Networks in Metagenomics
Sep 06, 2017
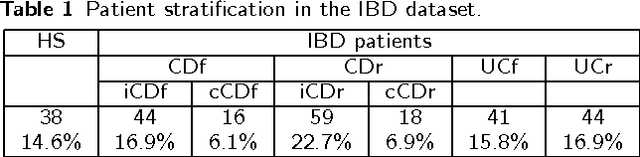

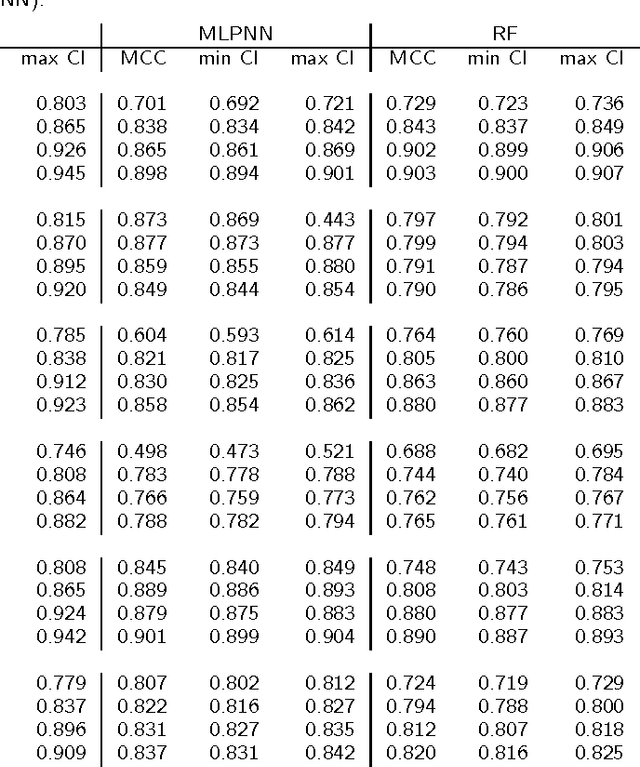
Abstract:Background: Convolutional Neural Networks can be effectively used only when data are endowed with an intrinsic concept of neighbourhood in the input space, as is the case of pixels in images. We introduce here Ph-CNN, a novel deep learning architecture for the classification of metagenomics data based on the Convolutional Neural Networks, with the patristic distance defined on the phylogenetic tree being used as the proximity measure. The patristic distance between variables is used together with a sparsified version of MultiDimensional Scaling to embed the phylogenetic tree in a Euclidean space. Results: Ph-CNN is tested with a domain adaptation approach on synthetic data and on a metagenomics collection of gut microbiota of 38 healthy subjects and 222 Inflammatory Bowel Disease patients, divided in 6 subclasses. Classification performance is promising when compared to classical algorithms like Support Vector Machines and Random Forest and a baseline fully connected neural network, e.g. the Multi-Layer Perceptron. Conclusion: Ph-CNN represents a novel deep learning approach for the classification of metagenomics data. Operatively, the algorithm has been implemented as a custom Keras layer taking care of passing to the following convolutional layer not only the data but also the ranked list of neighbourhood of each sample, thus mimicking the case of image data, transparently to the user. Keywords: Metagenomics; Deep learning; Convolutional Neural Networks; Phylogenetic trees
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge