Giuseppe Jurman
Deconver: A Deconvolutional Network for Medical Image Segmentation
Apr 01, 2025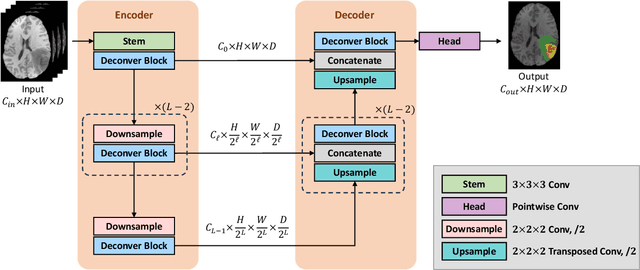
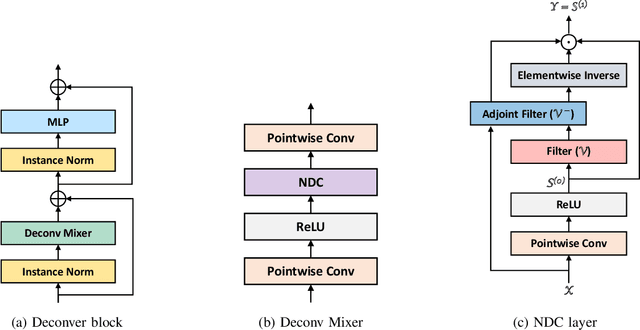
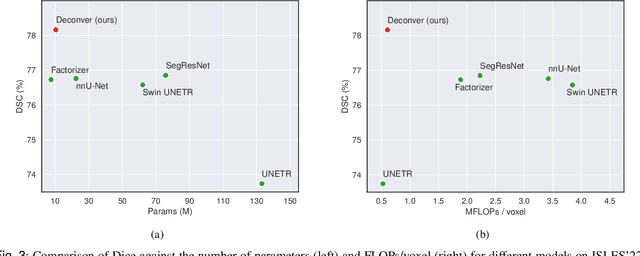
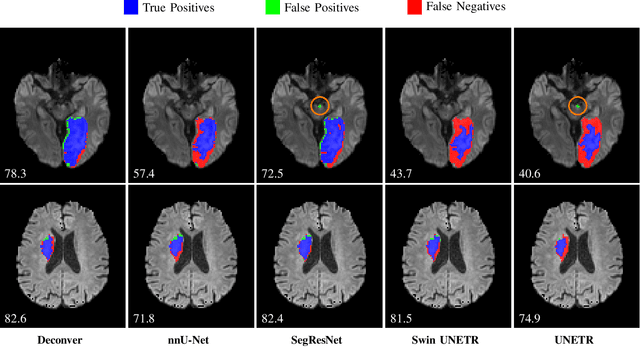
Abstract:While convolutional neural networks (CNNs) and vision transformers (ViTs) have advanced medical image segmentation, they face inherent limitations such as local receptive fields in CNNs and high computational complexity in ViTs. This paper introduces Deconver, a novel network that integrates traditional deconvolution techniques from image restoration as a core learnable component within a U-shaped architecture. Deconver replaces computationally expensive attention mechanisms with efficient nonnegative deconvolution (NDC) operations, enabling the restoration of high-frequency details while suppressing artifacts. Key innovations include a backpropagation-friendly NDC layer based on a provably monotonic update rule and a parameter-efficient design. Evaluated across four datasets (ISLES'22, BraTS'23, GlaS, FIVES) covering both 2D and 3D segmentation tasks, Deconver achieves state-of-the-art performance in Dice scores and Hausdorff distance while reducing computational costs (FLOPs) by up to 90% compared to leading baselines. By bridging traditional image restoration with deep learning, this work offers a practical solution for high-precision segmentation in resource-constrained clinical workflows. The project is available at https://github.com/pashtari/deconver.
Mitigating Health Data Poverty: Generative Approaches versus Resampling for Time-series Clinical Data
Oct 26, 2022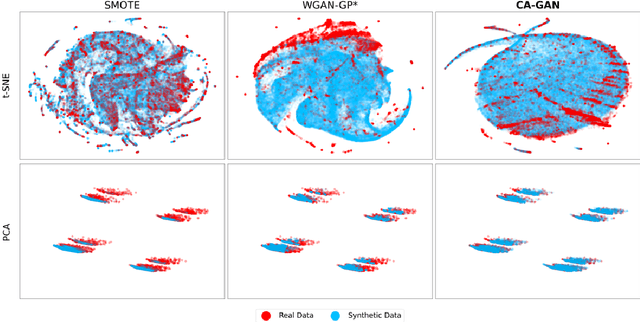
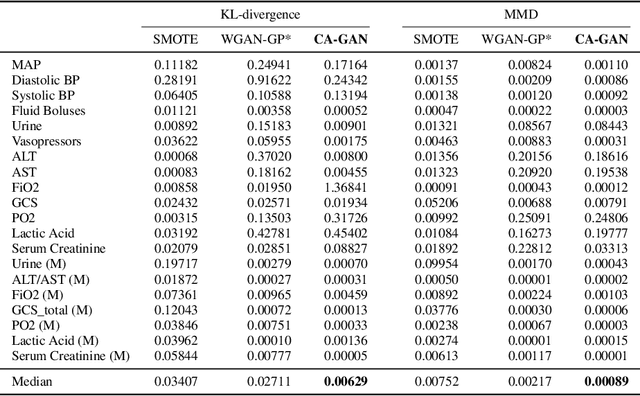
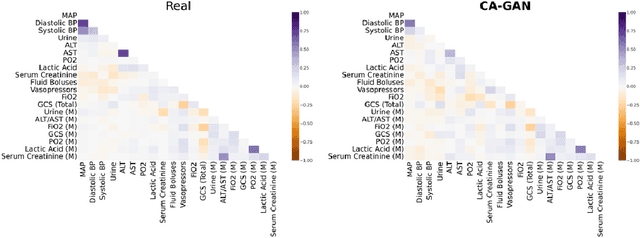
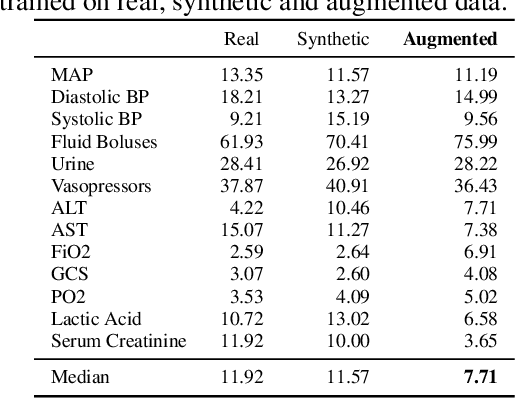
Abstract:Several approaches have been developed to mitigate algorithmic bias stemming from health data poverty, where minority groups are underrepresented in training datasets. Augmenting the minority class using resampling (such as SMOTE) is a widely used approach due to the simplicity of the algorithms. However, these algorithms decrease data variability and may introduce correlations between samples, giving rise to the use of generative approaches based on GAN. Generation of high-dimensional, time-series, authentic data that provides a wide distribution coverage of the real data, remains a challenging task for both resampling and GAN-based approaches. In this work we propose CA-GAN architecture that addresses some of the shortcomings of the current approaches, where we provide a detailed comparison with both SMOTE and WGAN-GP*, using a high-dimensional, time-series, real dataset of 3343 hypotensive Caucasian and Black patients. We show that our approach is better at both generating authentic data of the minority class and remaining within the original distribution of the real data.
MASS-UMAP: Fast and accurate analog ensemble search in weather radar archive
Oct 01, 2019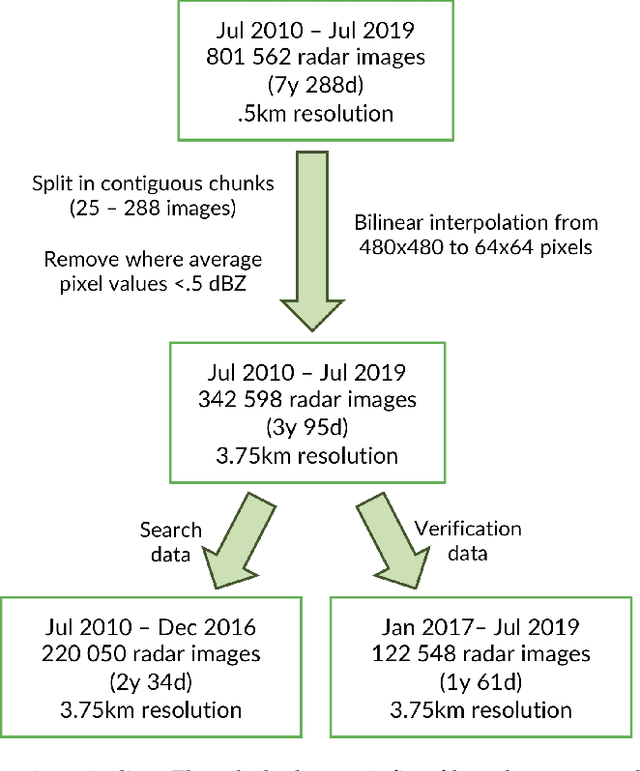
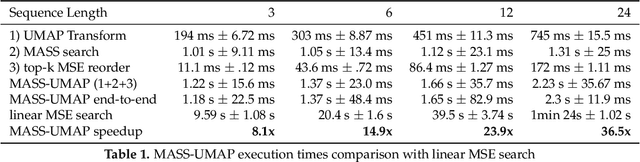
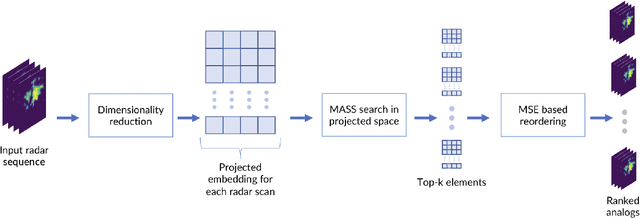
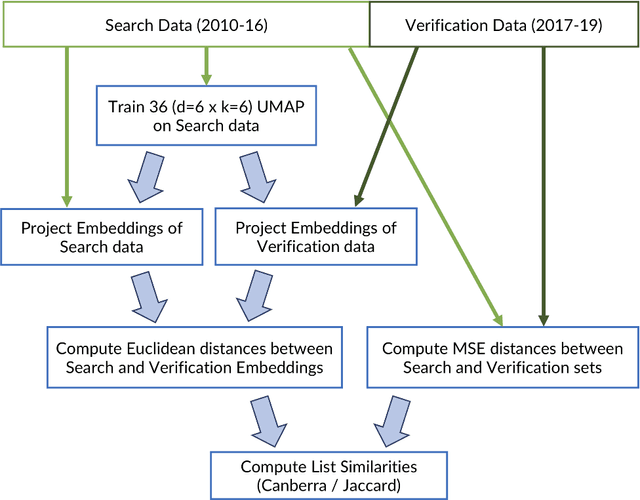
Abstract:The use of analogs - similar weather patterns - for weather forecasting and analysis is an established method in meteorology. The most challenging aspect of using this approach in the context of operational radar applications is to be able to perform a fast and accurate search for similar spatiotemporal precipitation patterns in a large archive of historical records. In this context, sequential pairwise search is too slow and computationally expensive. Here we propose an architecture to significantly speed-up spatiotemporal analog retrieval by combining nonlinear geometric dimensionality reduction (UMAP) with the fastest known Euclidean search algorithm for time series (MASS) to find radar analogs in constant time, independently of the desired temporal length to match and the number of extracted analogs. We compare UMAP with Principal component analysis (PCA) and show that UMAP outperforms PCA for spatial MSE analog search with proper settings. Moreover, we show that MASS is 20 times faster than brute force search on the UMAP embeddings space. We test the architecture on a real dataset and show that it enables precise and fast operational analog ensemble search through more than 2 years of radar archive in less than 5 seconds on a single workstation.
High Resolution Forecasting of Heat Waves impacts on Leaf Area Index by Multiscale Multitemporal Deep Learning
Sep 13, 2019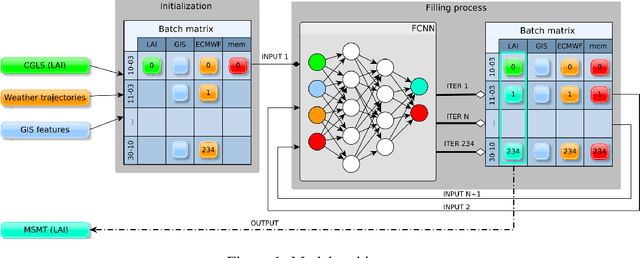

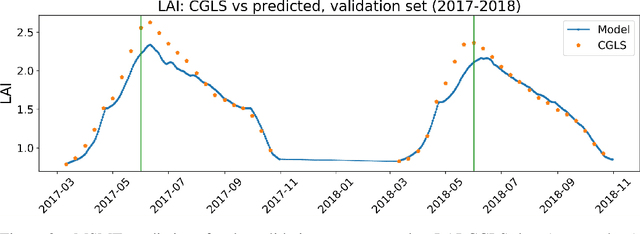
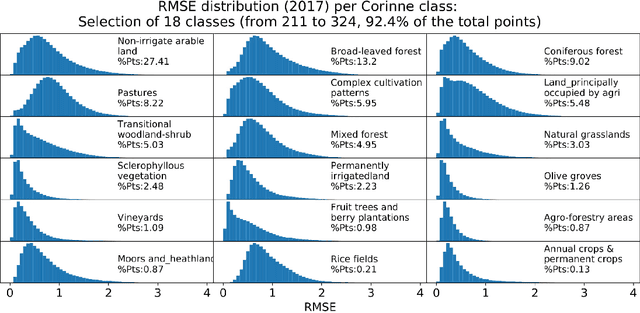
Abstract:Climate change impacts could cause progressive decrease of crop quality and yield, up to harvest failures. In particular, heat waves and other climate extremes can lead to localized food shortages and even threaten food security of communities worldwide. In this study, we apply a deep learning architecture for high resolution forecasting (300 m, 10 days) of the Leaf Area Index (LAI), whose dynamics has been widely used to model the growth phase of crops and impact of heat waves. LAI models can be computed at 0.1 degree spatial resolution with an auto regressive component adjusted with weather conditions, validated with remote sensing measurements. However model actionability is poor in regions of varying terrain morphology at this scale (about 8 km at the Alps latitude). Our deep learning model aims instead at forecasting LAI by training multiscale multitemporal (MSMT) data from the Copernicus Global Land Service (CGLS) project for all Europe at 300m resolution and medium-resolution historical weather data. Further, the deep learning model inputs integrate high-resolution land surface features, known to improve forecasts of agricultural productivity. The historical weather data are then replaced with forecast values to predict LAI values at 10 day horizon on Europe. We propose the MSMT model to develop a high resolution crop-specific warning system for mitigating damage due to heat waves and other extreme events.
A multiobjective deep learning approach for predictive classification in Neuroblastoma
Feb 22, 2018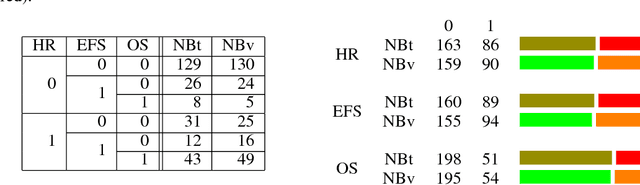
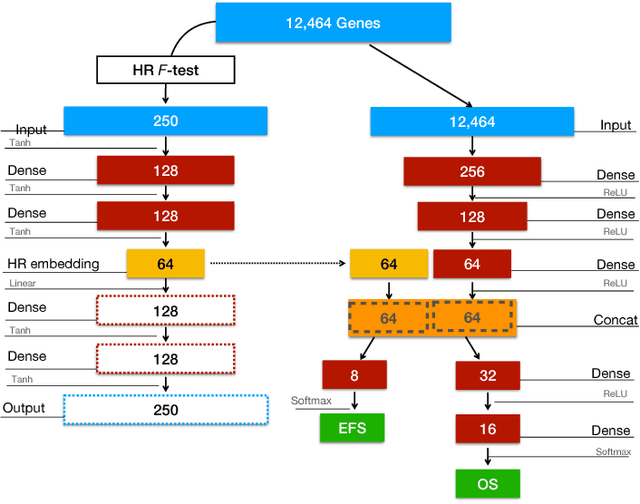
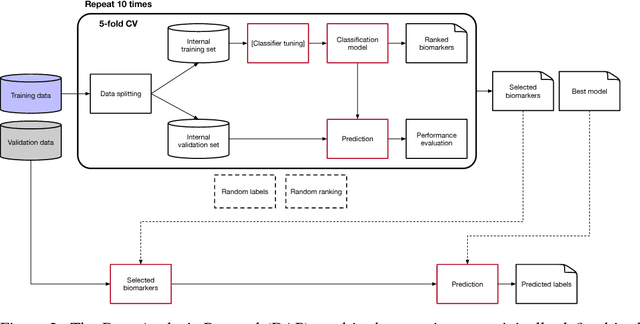
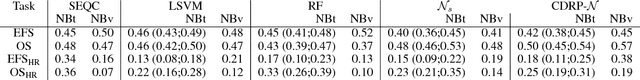
Abstract:Neuroblastoma is a strongly heterogeneous cancer with very diverse clinical courses that may vary from spontaneous regression to fatal progression; an accurate patient's risk estimation at diagnosis is essential to design appropriate tumor treatment strategies. Neuroblastoma is a paradigm disease where different diagnostic and prognostic endpoints should be predicted from common molecular and clinical information, with increasing complexity, as shown in the FDA MAQC-II study. Here we introduce the novel multiobjective deep learning architecture CDRP (Concatenated Diagnostic Relapse Prognostic) composed by 8 layers to obtain a combined diagnostic and prognostic prediction from high-throughput transcriptomics data. Two distinct loss functions are optimized for the Event Free Survival (EFS) and Overall Survival (OS) prognosis, respectively. We use the High-Risk (HR) diagnostic information as an additional input generated by an autoencoder embedding. The latter is used as network regulariser, based on a clinical algorithm commonly adopted for stratifying patients from cancer stage, age at insurgence of disease, and MYCN, the specific molecular marker. The architecture was applied to Illumina HiSeq2000 RNA-Seq for 498 neuroblastoma patients (176 at high risk) from the Sequencing Quality Control (SEQC) study, obtaining state-of-art on the diagnostic endpoint and improving prediction of prognosis over the HR cohort.
Convolutional neural networks for structured omics: OmicsCNN and the OmicsConv layer
Oct 16, 2017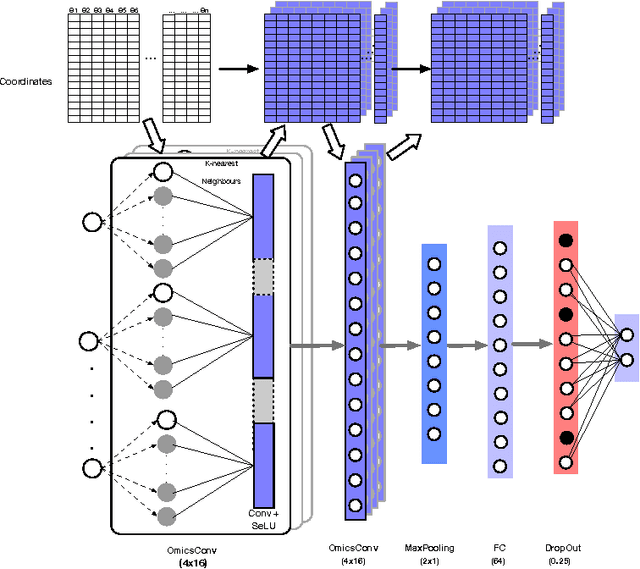

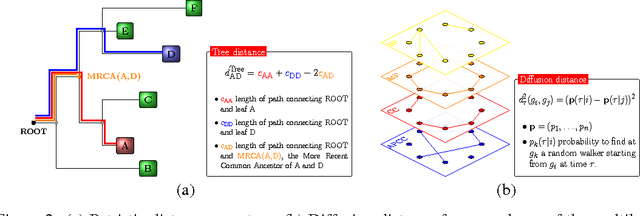
Abstract:Convolutional Neural Networks (CNNs) are a popular deep learning architecture widely applied in different domains, in particular in classifying over images, for which the concept of convolution with a filter comes naturally. Unfortunately, the requirement of a distance (or, at least, of a neighbourhood function) in the input feature space has so far prevented its direct use on data types such as omics data. However, a number of omics data are metrizable, i.e., they can be endowed with a metric structure, enabling to adopt a convolutional based deep learning framework, e.g., for prediction. We propose a generalized solution for CNNs on omics data, implemented through a dedicated Keras layer. In particular, for metagenomics data, a metric can be derived from the patristic distance on the phylogenetic tree. For transcriptomics data, we combine Gene Ontology semantic similarity and gene co-expression to define a distance; the function is defined through a multilayer network where 3 layers are defined by the GO mutual semantic similarity while the fourth one by gene co-expression. As a general tool, feature distance on omics data is enabled by OmicsConv, a novel Keras layer, obtaining OmicsCNN, a dedicated deep learning framework. Here we demonstrate OmicsCNN on gut microbiota sequencing data, for Inflammatory Bowel Disease (IBD) 16S data, first on synthetic data and then a metagenomics collection of gut microbiota of 222 IBD patients.
Deep Learning for Automatic Stereotypical Motor Movement Detection using Wearable Sensors in Autism Spectrum Disorders
Sep 14, 2017
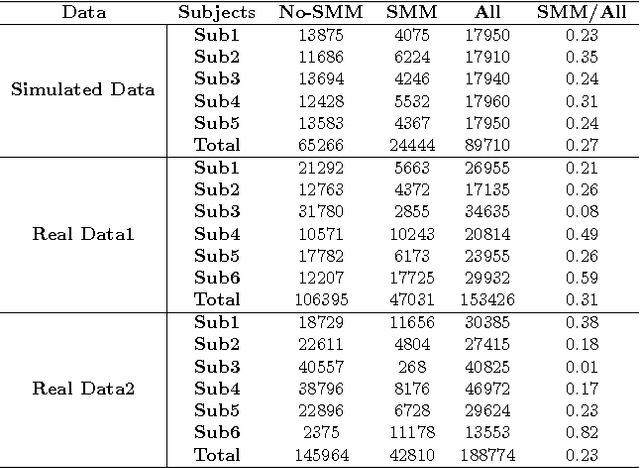
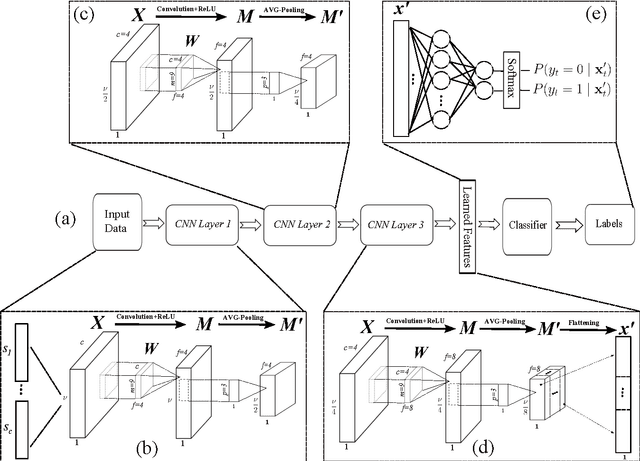
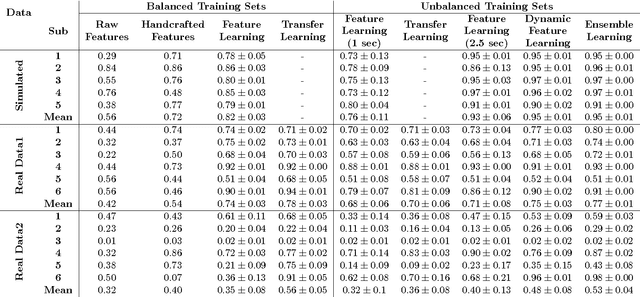
Abstract:Autism Spectrum Disorders are associated with atypical movements, of which stereotypical motor movements (SMMs) interfere with learning and social interaction. The automatic SMM detection using inertial measurement units (IMU) remains complex due to the strong intra and inter-subject variability, especially when handcrafted features are extracted from the signal. We propose a new application of the deep learning to facilitate automatic SMM detection using multi-axis IMUs. We use a convolutional neural network (CNN) to learn a discriminative feature space from raw data. We show how the CNN can be used for parameter transfer learning to enhance the detection rate on longitudinal data. We also combine the long short-term memory (LSTM) with CNN to model the temporal patterns in a sequence of multi-axis signals. Further, we employ ensemble learning to combine multiple LSTM learners into a more robust SMM detector. Our results show that: 1) feature learning outperforms handcrafted features; 2) parameter transfer learning is beneficial in longitudinal settings; 3) using LSTM to learn the temporal dynamic of signals enhances the detection rate especially for skewed training data; 4) an ensemble of LSTMs provides more accurate and stable detectors. These findings provide a significant step toward accurate SMM detection in real-time scenarios.
Phylogenetic Convolutional Neural Networks in Metagenomics
Sep 06, 2017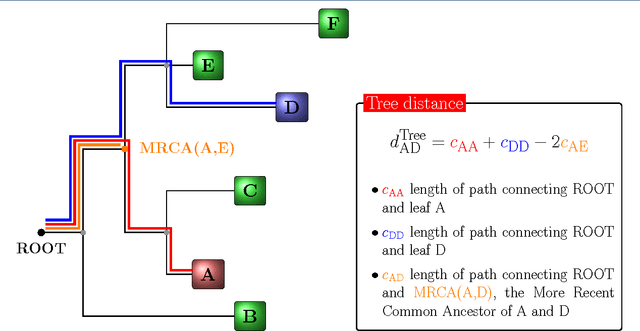
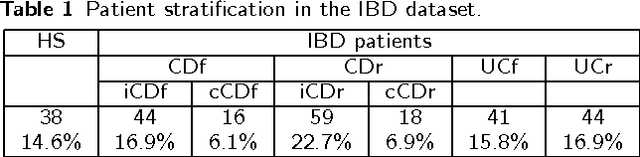

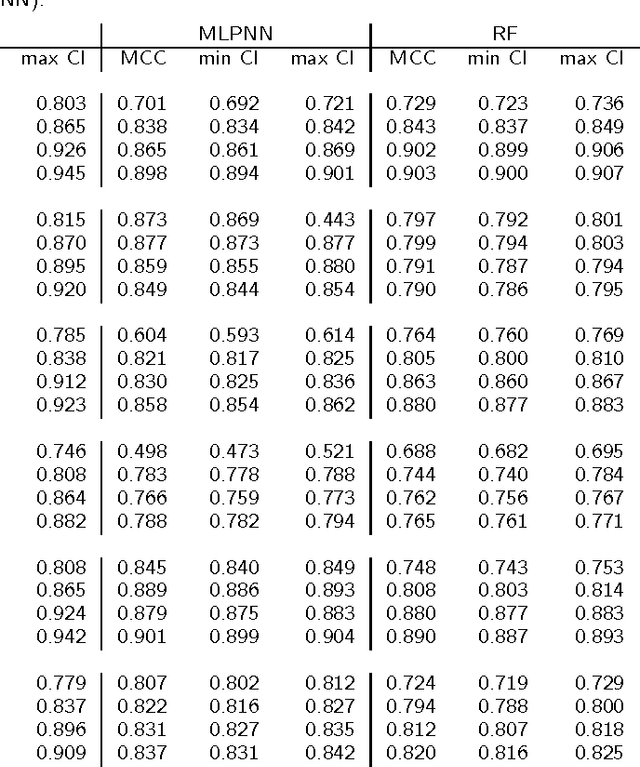
Abstract:Background: Convolutional Neural Networks can be effectively used only when data are endowed with an intrinsic concept of neighbourhood in the input space, as is the case of pixels in images. We introduce here Ph-CNN, a novel deep learning architecture for the classification of metagenomics data based on the Convolutional Neural Networks, with the patristic distance defined on the phylogenetic tree being used as the proximity measure. The patristic distance between variables is used together with a sparsified version of MultiDimensional Scaling to embed the phylogenetic tree in a Euclidean space. Results: Ph-CNN is tested with a domain adaptation approach on synthetic data and on a metagenomics collection of gut microbiota of 38 healthy subjects and 222 Inflammatory Bowel Disease patients, divided in 6 subclasses. Classification performance is promising when compared to classical algorithms like Support Vector Machines and Random Forest and a baseline fully connected neural network, e.g. the Multi-Layer Perceptron. Conclusion: Ph-CNN represents a novel deep learning approach for the classification of metagenomics data. Operatively, the algorithm has been implemented as a custom Keras layer taking care of passing to the following convolutional layer not only the data but also the ranked list of neighbourhood of each sample, thus mimicking the case of image data, transparently to the user. Keywords: Metagenomics; Deep learning; Convolutional Neural Networks; Phylogenetic trees
Towards meaningful physics from generative models
May 26, 2017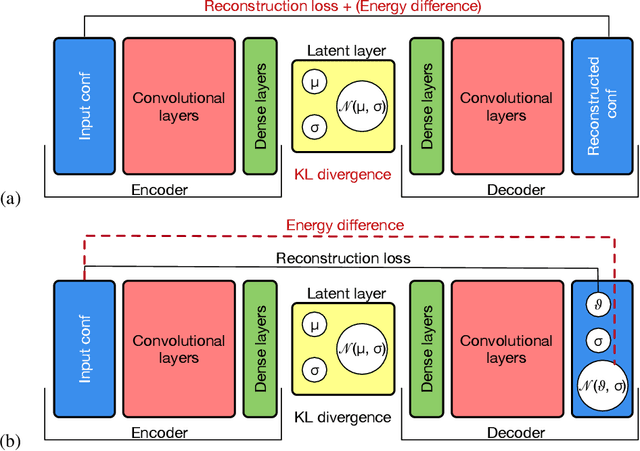
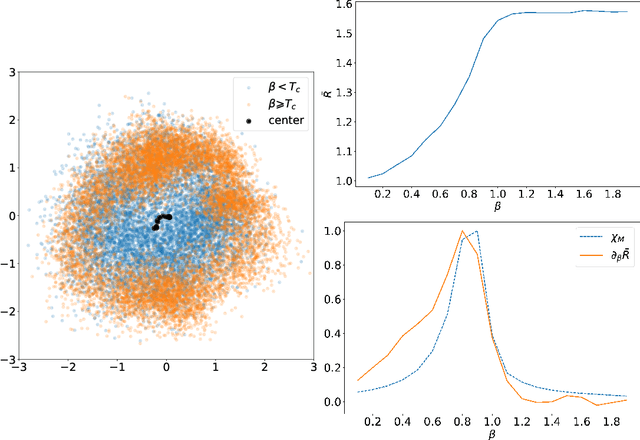
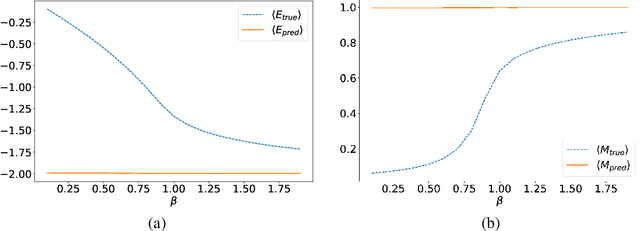
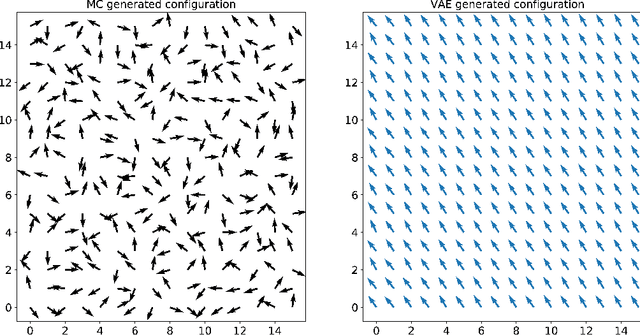
Abstract:In several physical systems, important properties characterizing the system itself are theoretically related with specific degrees of freedom. Although standard Monte Carlo simulations provide an effective tool to accurately reconstruct the physical configurations of the system, they are unable to isolate the different contributions corresponding to different degrees of freedom. Here we show that unsupervised deep learning can become a valid support to MC simulation, coupling useful insights in the phases detection task with good reconstruction performance. As a testbed we consider the 2D XY model, showing that a deep neural network based on variational autoencoders can detect the continuous Kosterlitz-Thouless (KT) transitions, and that, if endowed with the appropriate constrains, they generate configurations with meaningful physical content.
Convolutional Neural Network for Stereotypical Motor Movement Detection in Autism
Jun 07, 2016
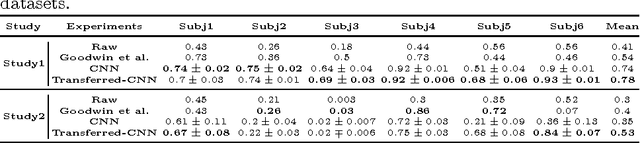

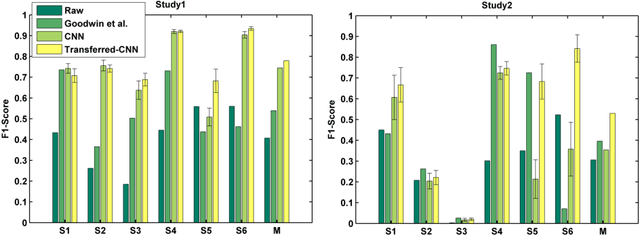
Abstract:Autism Spectrum Disorders (ASDs) are often associated with specific atypical postural or motor behaviors, of which Stereotypical Motor Movements (SMMs) have a specific visibility. While the identification and the quantification of SMM patterns remain complex, its automation would provide support to accurate tuning of the intervention in the therapy of autism. Therefore, it is essential to develop automatic SMM detection systems in a real world setting, taking care of strong inter-subject and intra-subject variability. Wireless accelerometer sensing technology can provide a valid infrastructure for real-time SMM detection, however such variability remains a problem also for machine learning methods, in particular whenever handcrafted features extracted from accelerometer signal are considered. Here, we propose to employ the deep learning paradigm in order to learn discriminating features from multi-sensor accelerometer signals. Our results provide preliminary evidence that feature learning and transfer learning embedded in the deep architecture achieve higher accurate SMM detectors in longitudinal scenarios.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge