Sabine Van Huffel
Deconver: A Deconvolutional Network for Medical Image Segmentation
Apr 01, 2025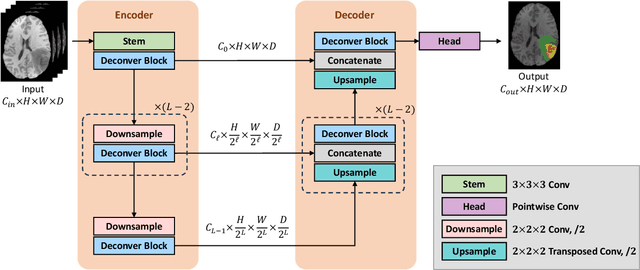
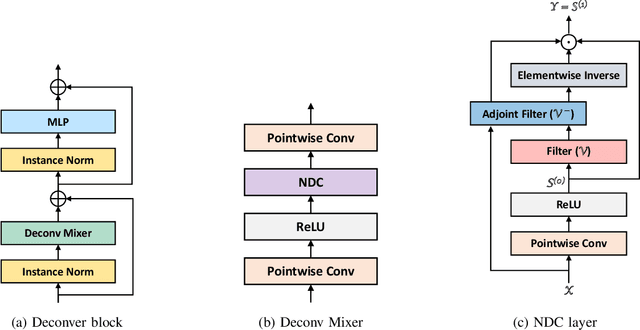
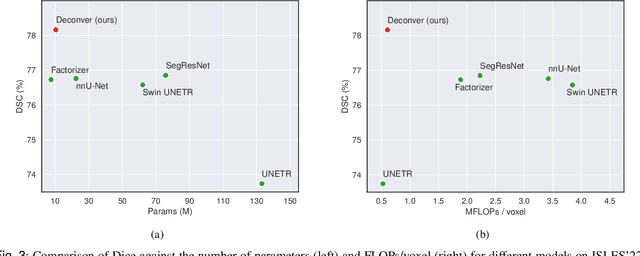
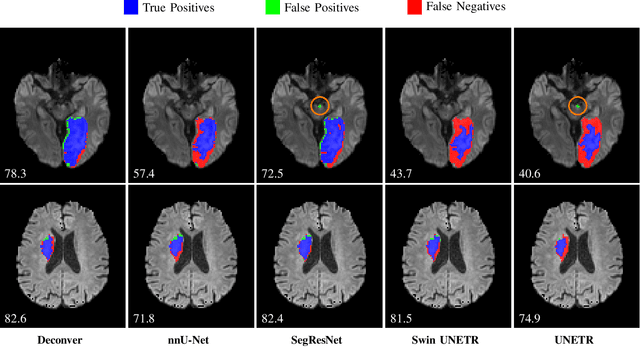
Abstract:While convolutional neural networks (CNNs) and vision transformers (ViTs) have advanced medical image segmentation, they face inherent limitations such as local receptive fields in CNNs and high computational complexity in ViTs. This paper introduces Deconver, a novel network that integrates traditional deconvolution techniques from image restoration as a core learnable component within a U-shaped architecture. Deconver replaces computationally expensive attention mechanisms with efficient nonnegative deconvolution (NDC) operations, enabling the restoration of high-frequency details while suppressing artifacts. Key innovations include a backpropagation-friendly NDC layer based on a provably monotonic update rule and a parameter-efficient design. Evaluated across four datasets (ISLES'22, BraTS'23, GlaS, FIVES) covering both 2D and 3D segmentation tasks, Deconver achieves state-of-the-art performance in Dice scores and Hausdorff distance while reducing computational costs (FLOPs) by up to 90% compared to leading baselines. By bridging traditional image restoration with deep learning, this work offers a practical solution for high-precision segmentation in resource-constrained clinical workflows. The project is available at https://github.com/pashtari/deconver.
Quantization-free Lossy Image Compression Using Integer Matrix Factorization
Aug 22, 2024



Abstract:Lossy image compression is essential for efficient transmission and storage. Traditional compression methods mainly rely on discrete cosine transform (DCT) or singular value decomposition (SVD), both of which represent image data in continuous domains and therefore necessitate carefully designed quantizers. Notably, SVD-based methods are more sensitive to quantization errors than DCT-based methods like JPEG. To address this issue, we introduce a variant of integer matrix factorization (IMF) to develop a novel quantization-free lossy image compression method. IMF provides a low-rank representation of the image data as a product of two smaller factor matrices with bounded integer elements, thereby eliminating the need for quantization. We propose an efficient, provably convergent iterative algorithm for IMF using a block coordinate descent (BCD) scheme, with subproblems having closed-form solutions. Our experiments on the Kodak and CLIC 2024 datasets demonstrate that our IMF compression method consistently outperforms JPEG at low bit rates below 0.25 bits per pixel (bpp) and remains comparable at higher bit rates. We also assessed our method's capability to preserve visual semantics by evaluating an ImageNet pre-trained classifier on compressed images. Remarkably, our method improved top-1 accuracy by over 5 percentage points compared to JPEG at bit rates under 0.25 bpp. The project is available at https://github.com/pashtari/lrf .
A Robust Ensemble Algorithm for Ischemic Stroke Lesion Segmentation: Generalizability and Clinical Utility Beyond the ISLES Challenge
Apr 03, 2024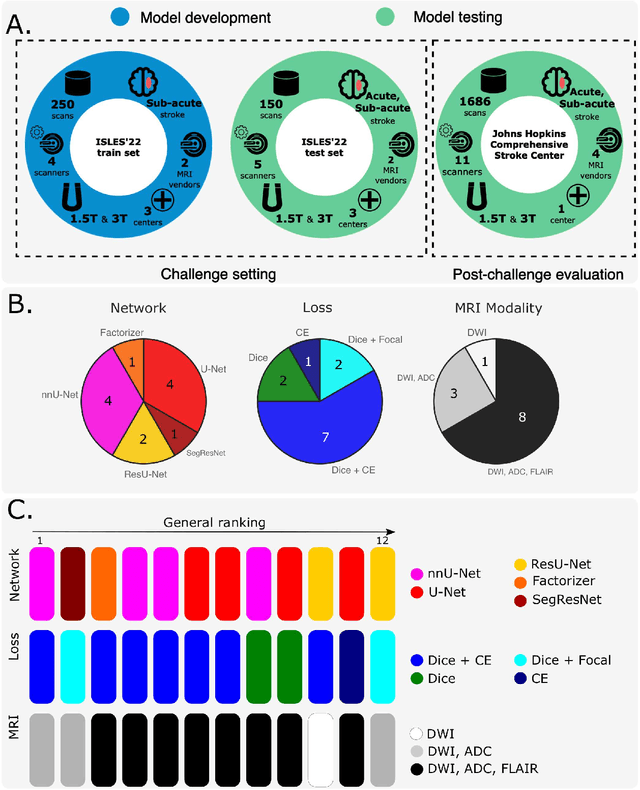
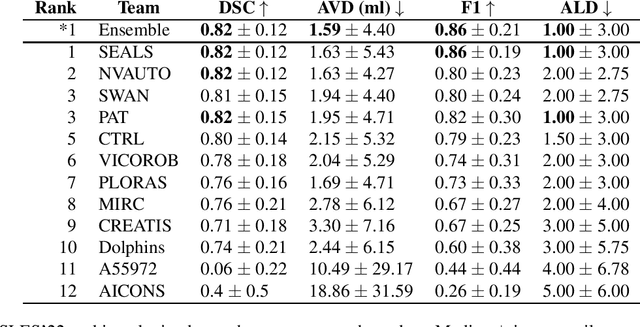
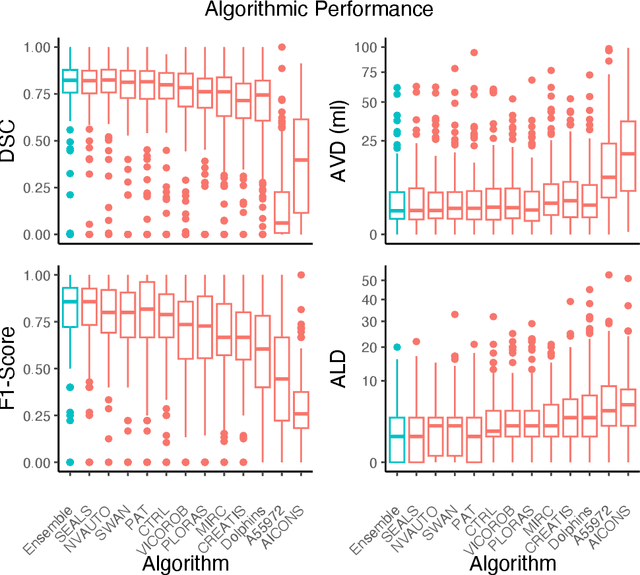
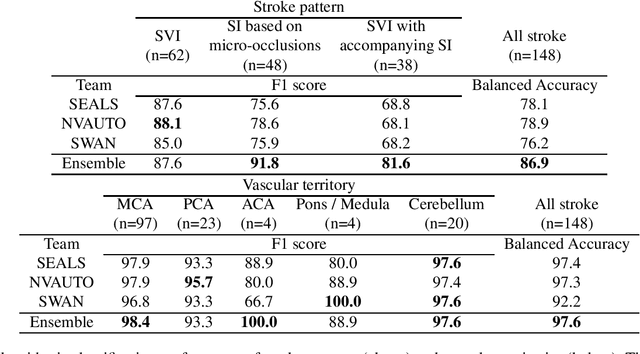
Abstract:Diffusion-weighted MRI (DWI) is essential for stroke diagnosis, treatment decisions, and prognosis. However, image and disease variability hinder the development of generalizable AI algorithms with clinical value. We address this gap by presenting a novel ensemble algorithm derived from the 2022 Ischemic Stroke Lesion Segmentation (ISLES) challenge. ISLES'22 provided 400 patient scans with ischemic stroke from various medical centers, facilitating the development of a wide range of cutting-edge segmentation algorithms by the research community. Through collaboration with leading teams, we combined top-performing algorithms into an ensemble model that overcomes the limitations of individual solutions. Our ensemble model achieved superior ischemic lesion detection and segmentation accuracy on our internal test set compared to individual algorithms. This accuracy generalized well across diverse image and disease variables. Furthermore, the model excelled in extracting clinical biomarkers. Notably, in a Turing-like test, neuroradiologists consistently preferred the algorithm's segmentations over manual expert efforts, highlighting increased comprehensiveness and precision. Validation using a real-world external dataset (N=1686) confirmed the model's generalizability. The algorithm's outputs also demonstrated strong correlations with clinical scores (admission NIHSS and 90-day mRS) on par with or exceeding expert-derived results, underlining its clinical relevance. This study offers two key findings. First, we present an ensemble algorithm (https://github.com/Tabrisrei/ISLES22_Ensemble) that detects and segments ischemic stroke lesions on DWI across diverse scenarios on par with expert (neuro)radiologists. Second, we show the potential for biomedical challenge outputs to extend beyond the challenge's initial objectives, demonstrating their real-world clinical applicability.
Machine Learning to Support Triage of Children at Risk for Epileptic Seizures in the Pediatric Intensive Care Unit
May 11, 2022



Abstract:Objective: Epileptic seizures are relatively common in critically-ill children admitted to the pediatric intensive care unit (PICU) and thus serve as an important target for identification and treatment. Most of these seizures have no discernible clinical manifestation but still have a significant impact on morbidity and mortality. Children that are deemed at risk for seizures within the PICU are monitored using continuous-electroencephalogram (cEEG). cEEG monitoring cost is considerable and as the number of available machines is always limited, clinicians need to resort to triaging patients according to perceived risk in order to allocate resources. This research aims to develop a computer aided tool to improve seizures risk assessment in critically-ill children, using an ubiquitously recorded signal in the PICU, namely the electrocardiogram (ECG). Approach: A novel data-driven model was developed at a patient-level approach, based on features extracted from the first hour of ECG recording and the clinical data of the patient. Main results: The most predictive features were the age of the patient, the brain injury as coma etiology and the QRS area. For patients without any prior clinical data, using one hour of ECG recording, the classification performance of the random forest classifier reached an area under the receiver operating characteristic curve (AUROC) score of 0.84. When combining ECG features with the patients clinical history, the AUROC reached 0.87. Significance: Taking a real clinical scenario, we estimated that our clinical decision support triage tool can improve the positive predictive value by more than 59% over the clinical standard.
Factorizer: A Scalable Interpretable Approach to Context Modeling for Medical Image Segmentation
Feb 28, 2022
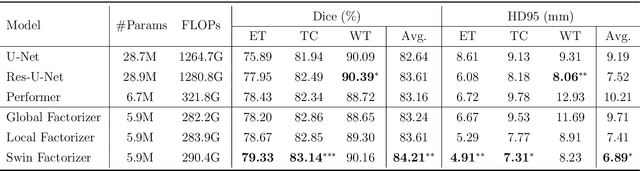


Abstract:Convolutional Neural Networks (CNNs) with U-shaped architectures have dominated medical image segmentation, which is crucial for various clinical purposes. However, the inherent locality of convolution makes CNNs fail to fully exploit global context, essential for better recognition of some structures, e.g., brain lesions. Transformers have recently proved promising performance on vision tasks, including semantic segmentation, mainly due to their capability of modeling long-range dependencies. Nevertheless, the quadratic complexity of attention makes existing Transformer-based models use self-attention layers only after somehow reducing the image resolution, which limits the ability to capture global contexts present at higher resolutions. Therefore, this work introduces a family of models, dubbed Factorizer, which leverages the power of low-rank matrix factorization for constructing an end-to-end segmentation model. Specifically, we propose a linearly scalable approach to context modeling, formulating Nonnegative Matrix Factorization (NMF) as a differentiable layer integrated into a U-shaped architecture. The shifted window technique is also utilized in combination with NMF to effectively aggregate local information. Factorizers compete favorably with CNNs and Transformers in terms of accuracy, scalability, and interpretability, achieving state-of-the-art results on the BraTS dataset for brain tumor segmentation, with Dice scores of 79.33%, 83.14%, and 90.16% for enhancing tumor, tumor core, and whole tumor, respectively. Highly meaningful NMF components give an additional interpretability advantage to Factorizers over CNNs and Transformers. Moreover, our ablation studies reveal a distinctive feature of Factorizers that enables a significant speed-up in inference for a trained Factorizer without any extra steps and without sacrificing much accuracy.
Early soft and flexible fusion of EEG and fMRI via tensor decompositions
May 12, 2020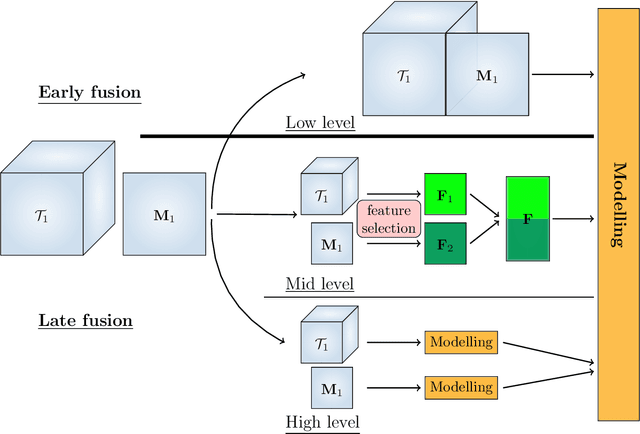
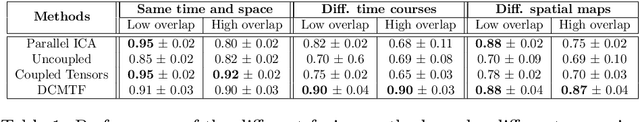
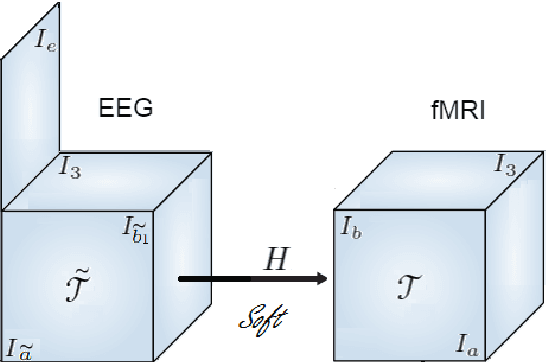
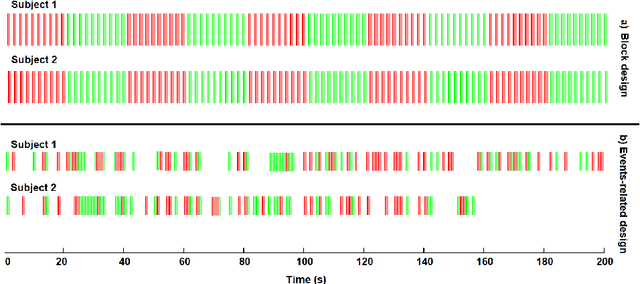
Abstract:Data fusion refers to the joint analysis of multiple datasets which provide complementary views of the same task. In this preprint, the problem of jointly analyzing electroencephalography (EEG) and functional Magnetic Resonance Imaging (fMRI) data is considered. Jointly analyzing EEG and fMRI measurements is highly beneficial for studying brain function because these modalities have complementary spatiotemporal resolution: EEG offers good temporal resolution while fMRI is better in its spatial resolution. The fusion methods reported so far ignore the underlying multi-way nature of the data in at least one of the modalities and/or rely on very strong assumptions about the relation of the two datasets. In this preprint, these two points are addressed by adopting for the first time tensor models in the two modalities while also exploring double coupled tensor decompositions and by following soft and flexible coupling approaches to implement the multi-modal analysis. To cope with the Event Related Potential (ERP) variability in EEG, the PARAFAC2 model is adopted. The results obtained are compared against those of parallel Independent Component Analysis (ICA) and hard coupling alternatives in both simulated and real data. Our results confirm the superiority of tensorial methods over methods based on ICA. In scenarios that do not meet the assumptions underlying hard coupling, the advantage of soft and flexible coupled decompositions is clearly demonstrated.
Augmenting interictal mapping with neurovascular coupling biomarkers by structured factorization of epileptic EEG and fMRI data
Apr 29, 2020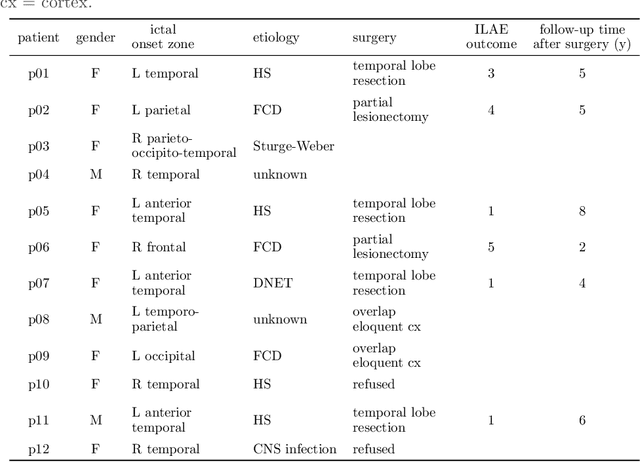



Abstract:EEG-correlated fMRI analysis is widely used to detect regional blood oxygen level dependent fluctuations that are significantly synchronized to interictal epileptic discharges, which can provide evidence for localizing the ictal onset zone. However, such an asymmetrical, mass-univariate approach cannot capture the inherent, higher order structure in the EEG data, nor multivariate relations in the fMRI data, and it is nontrivial to accurately handle varying neurovascular coupling over patients and brain regions. We aim to overcome these drawbacks in a data-driven manner by means of a novel structured matrix-tensor factorization: the single-subject EEG data (represented as a third-order spectrogram tensor) and fMRI data (represented as a spatiotemporal BOLD signal matrix) are jointly decomposed into a superposition of several sources, characterized by space-time-frequency profiles. In the shared temporal mode, Toeplitz-structured factors account for a spatially specific, neurovascular `bridge' between the EEG and fMRI temporal fluctuations, capturing the hemodynamic response's variability over brain regions. We show that the extracted source signatures provide a sensitive localization of the ictal onset zone, and, moreover, that complementary localizing information can be derived from the spatial variation of the hemodynamic response. Hence, this multivariate, multimodal factorization provides two useful sets of EEG-fMRI biomarkers, which can inform the presurgical evaluation of epilepsy. We make all code required to perform the computations available.
Compressed Sensing of Multi-Channel EEG Signals: The Simultaneous Cosparsity and Low Rank Optimization
Jun 29, 2015


Abstract:Goal: This paper deals with the problems that some EEG signals have no good sparse representation and single channel processing is not computationally efficient in compressed sensing of multi-channel EEG signals. Methods: An optimization model with L0 norm and Schatten-0 norm is proposed to enforce cosparsity and low rank structures in the reconstructed multi-channel EEG signals. Both convex relaxation and global consensus optimization with alternating direction method of multipliers are used to compute the optimization model. Results: The performance of multi-channel EEG signal reconstruction is improved in term of both accuracy and computational complexity. Conclusion: The proposed method is a better candidate than previous sparse signal recovery methods for compressed sensing of EEG signals. Significance: The proposed method enables successful compressed sensing of EEG signals even when the signals have no good sparse representation. Using compressed sensing would much reduce the power consumption of wireless EEG system.
Multi-Sparse Signal Recovery for Compressive Sensing
Jun 04, 2012



Abstract:Signal recovery is one of the key techniques of Compressive sensing (CS). It reconstructs the original signal from the linear sub-Nyquist measurements. Classical methods exploit the sparsity in one domain to formulate the L0 norm optimization. Recent investigation shows that some signals are sparse in multiple domains. To further improve the signal reconstruction performance, we can exploit this multi-sparsity to generate a new convex programming model. The latter is formulated with multiple sparsity constraints in multiple domains and the linear measurement fitting constraint. It improves signal recovery performance by additional a priori information. Since some EMG signals exhibit sparsity both in time and frequency domains, we take them as example in numerical experiments. Results show that the newly proposed method achieves better performance for multi-sparse signals.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge