Maarten De Vos
University of Oxford, Institute of Biomedical Engineering, Dept. Engineering Sciences, Oxford, UK
SeizeIT2: Wearable Dataset Of Patients With Focal Epilepsy
Feb 03, 2025



Abstract:The increasing technological advancements towards miniaturized physiological measuring devices have enabled continuous monitoring of epileptic patients outside of specialized environments. The large amounts of data that can be recorded with such devices holds significant potential for developing automated seizure detection frameworks. In this work, we present SeizeIT2, the first open dataset of wearable data recorded in patients with focal epilepsy. The dataset comprises more than 11,000 hours of multimodal data, including behind-the-ear electroencephalography, electrocardiography, electromyography and movement (accelerometer and gyroscope) data. The dataset contains 886 focal seizures recorded from 125 patients across five different European Epileptic Monitoring Centers. We present a suggestive training/validation split to propel the development of AI methodologies for seizure detection, as well as two benchmark approaches and evaluation metrics. The dataset can be accessed on OpenNeuro and is stored in Brain Imaging Data Structure (BIDS) format.
Multimodal Fusion Balancing Through Game-Theoretic Regularization
Nov 11, 2024



Abstract:Multimodal learning can complete the picture of information extraction by uncovering key dependencies between data sources. However, current systems fail to fully leverage multiple modalities for optimal performance. This has been attributed to modality competition, where modalities strive for training resources, leaving some underoptimized. We show that current balancing methods struggle to train multimodal models that surpass even simple baselines, such as ensembles. This raises the question: how can we ensure that all modalities in multimodal training are sufficiently trained, and that learning from new modalities consistently improves performance? This paper proposes the Multimodal Competition Regularizer (MCR), a new loss component inspired by mutual information (MI) decomposition designed to prevent the adverse effects of competition in multimodal training. Our key contributions are: 1) Introducing game-theoretic principles in multimodal learning, where each modality acts as a player competing to maximize its influence on the final outcome, enabling automatic balancing of the MI terms. 2) Refining lower and upper bounds for each MI term to enhance the extraction of task-relevant unique and shared information across modalities. 3) Suggesting latent space permutations for conditional MI estimation, significantly improving computational efficiency. MCR outperforms all previously suggested training strategies and is the first to consistently improve multimodal learning beyond the ensemble baseline, clearly demonstrating that combining modalities leads to significant performance gains on both synthetic and large real-world datasets.
MixNet: Joining Force of Classical and Modern Approaches Toward the Comprehensive Pipeline in Motor Imagery EEG Classification
Sep 06, 2024



Abstract:Recent advances in deep learning (DL) have significantly impacted motor imagery (MI)-based brain-computer interface (BCI) systems, enhancing the decoding of electroencephalography (EEG) signals. However, most studies struggle to identify discriminative patterns across subjects during MI tasks, limiting MI classification performance. In this article, we propose MixNet, a novel classification framework designed to overcome this limitation by utilizing spectral-spatial signals from MI data, along with a multitask learning architecture named MIN2Net, for classification. Here, the spectral-spatial signals are generated using the filter-bank common spatial patterns (FBCSPs) method on MI data. Since the multitask learning architecture is used for the classification task, the learning in each task may exhibit different generalization rates and potential overfitting across tasks. To address this issue, we implement adaptive gradient blending, simultaneously regulating multiple loss weights and adjusting the learning pace for each task based on its generalization/overfitting tendencies. Experimental results on six benchmark data sets of different data sizes demonstrate that MixNet consistently outperforms all state-of-the-art algorithms in subject-dependent and -independent settings. Finally, the low-density EEG MI classification results show that MixNet outperforms all state-of-the-art algorithms, offering promising implications for Internet of Thing (IoT) applications, such as lightweight and portable EEG wearable devices based on low-density montages.
* Supplementary materials and source codes are available on-line at https://github.com/Max-Phairot-A/MixNet
Improving Multimodal Learning with Multi-Loss Gradient Modulation
May 13, 2024



Abstract:Learning from multiple modalities, such as audio and video, offers opportunities for leveraging complementary information, enhancing robustness, and improving contextual understanding and performance. However, combining such modalities presents challenges, especially when modalities differ in data structure, predictive contribution, and the complexity of their learning processes. It has been observed that one modality can potentially dominate the learning process, hindering the effective utilization of information from other modalities and leading to sub-optimal model performance. To address this issue the vast majority of previous works suggest to assess the unimodal contributions and dynamically adjust the training to equalize them. We improve upon previous work by introducing a multi-loss objective and further refining the balancing process, allowing it to dynamically adjust the learning pace of each modality in both directions, acceleration and deceleration, with the ability to phase out balancing effects upon convergence. We achieve superior results across three audio-video datasets: on CREMA-D, models with ResNet backbone encoders surpass the previous best by 1.9% to 12.4%, and Conformer backbone models deliver improvements ranging from 2.8% to 14.1% across different fusion methods. On AVE, improvements range from 2.7% to 7.7%, while on UCF101, gains reach up to 6.1%.
Multimodal wearable EEG, EMG and accelerometry measurements improve the accuracy of tonic-clonic seizure detection in-hospital
Mar 19, 2024



Abstract:Objective: Most current wearable tonic-clonic seizure (TCS) detection systems are based on extra-cerebral signals, such as electromyography (EMG) or accelerometry (ACC). Although many of these devices show good sensitivity in seizure detection, their false positive rates (FPR) are still relatively high. Wearable EEG may improve performance; however, studies investigating this remain scarce. This paper aims 1) to investigate the possibility of detecting TCSs with a behind-the-ear, two-channel wearable EEG, and 2) to evaluate the added value of wearable EEG to other non-EEG modalities in multimodal TCS detection. Method: We included 27 participants with a total of 44 TCSs from the European multicenter study SeizeIT2. The multimodal wearable detection system Sensor Dot (Byteflies) was used to measure two-channel, behind-the-ear EEG, EMG, electrocardiography (ECG), ACC and gyroscope (GYR). First, we evaluated automatic unimodal detection of TCSs, using performance metrics such as sensitivity, precision, FPR and F1-score. Secondly, we fused the different modalities and again assessed performance. Algorithm-labeled segments were then provided to a neurologist and a wearable data expert, who reviewed and annotated the true positive TCSs, and discarded false positives (FPs). Results: Wearable EEG outperformed the other modalities in unimodal TCS detection by achieving a sensitivity of 100.0% and a FPR of 10.3/24h (compared to 97.7% sensitivity and 30.9/24h FPR for EMG; 95.5% sensitivity and 13.9 FPR for ACC). The combination of wearable EEG and EMG achieved overall the most clinically useful performance in offline TCS detection with a sensitivity of 97.7%, a FPR of 0.4/24 h, a precision of 43.0%, and a F1-score of 59.7%. Subsequent visual review of the automated detections resulted in maximal sensitivity and zero FPs.
Increasing Performance And Sample Efficiency With Model-agnostic Interactive Feature Attributions
Jun 28, 2023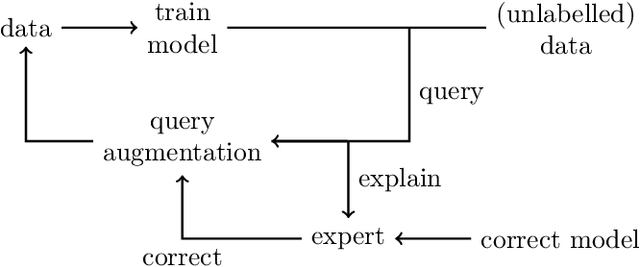
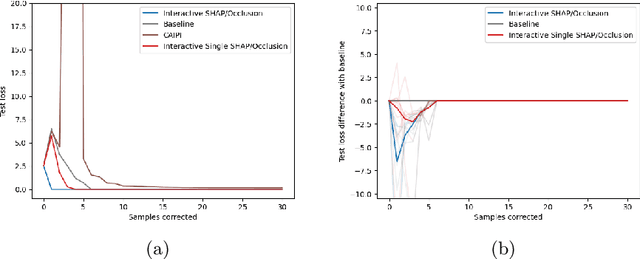
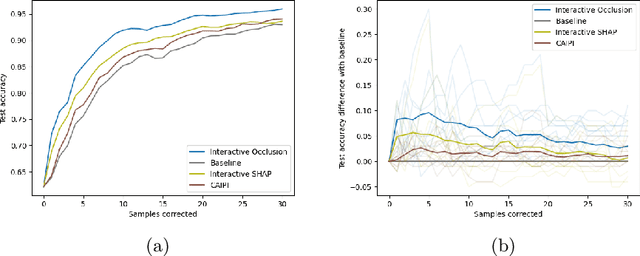
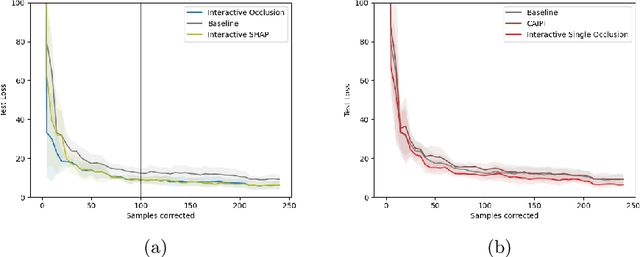
Abstract:Model-agnostic feature attributions can provide local insights in complex ML models. If the explanation is correct, a domain expert can validate and trust the model's decision. However, if it contradicts the expert's knowledge, related work only corrects irrelevant features to improve the model. To allow for unlimited interaction, in this paper we provide model-agnostic implementations for two popular explanation methods (Occlusion and Shapley values) to enforce entirely different attributions in the complex model. For a particular set of samples, we use the corrected feature attributions to generate extra local data, which is used to retrain the model to have the right explanation for the samples. Through simulated and real data experiments on a variety of models we show how our proposed approach can significantly improve the model's performance only by augmenting its training dataset based on corrected explanations. Adding our interactive explanations to active learning settings increases the sample efficiency significantly and outperforms existing explanatory interactive strategies. Additionally we explore how a domain expert can provide feature attributions which are sufficiently correct to improve the model.
Explaining the Model and Feature Dependencies by Decomposition of the Shapley Value
Jun 19, 2023Abstract:Shapley values have become one of the go-to methods to explain complex models to end-users. They provide a model agnostic post-hoc explanation with foundations in game theory: what is the worth of a player (in machine learning, a feature value) in the objective function (the output of the complex machine learning model). One downside is that they always require outputs of the model when some features are missing. These are usually computed by taking the expectation over the missing features. This however introduces a non-trivial choice: do we condition on the unknown features or not? In this paper we examine this question and claim that they represent two different explanations which are valid for different end-users: one that explains the model and one that explains the model combined with the feature dependencies in the data. We propose a new algorithmic approach to combine both explanations, removing the burden of choice and enhancing the explanatory power of Shapley values, and show that it achieves intuitive results on simple problems. We apply our method to two real-world datasets and discuss the explanations. Finally, we demonstrate how our method is either equivalent or superior to state-to-of-art Shapley value implementations while simultaneously allowing for increased insight into the model-data structure.
U-PASS: an Uncertainty-guided deep learning Pipeline for Automated Sleep Staging
Jun 07, 2023



Abstract:As machine learning becomes increasingly prevalent in critical fields such as healthcare, ensuring the safety and reliability of machine learning systems becomes paramount. A key component of reliability is the ability to estimate uncertainty, which enables the identification of areas of high and low confidence and helps to minimize the risk of error. In this study, we propose a machine learning pipeline called U-PASS tailored for clinical applications that incorporates uncertainty estimation at every stage of the process, including data acquisition, training, and model deployment. The training process is divided into a supervised pre-training step and a semi-supervised finetuning step. We apply our uncertainty-guided deep learning pipeline to the challenging problem of sleep staging and demonstrate that it systematically improves performance at every stage. By optimizing the training dataset, actively seeking informative samples, and deferring the most uncertain samples to an expert, we achieve an expert-level accuracy of 85% on a challenging clinical dataset of elderly sleep apnea patients, representing a significant improvement over the baseline accuracy of 75%. U-PASS represents a promising approach to incorporating uncertainty estimation into machine learning pipelines, thereby improving their reliability and unlocking their potential in clinical settings.
CoRe-Sleep: A Multimodal Fusion Framework for Time Series Robust to Imperfect Modalities
Mar 27, 2023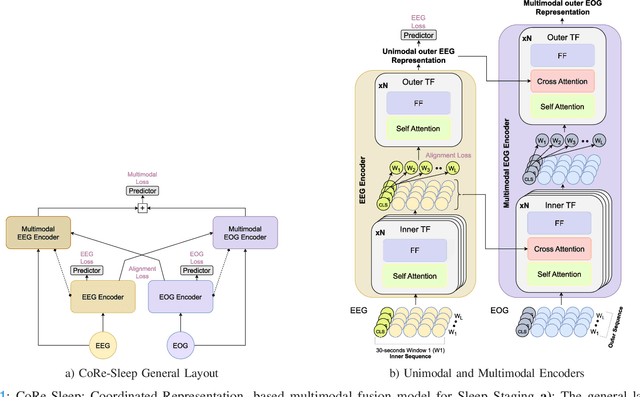
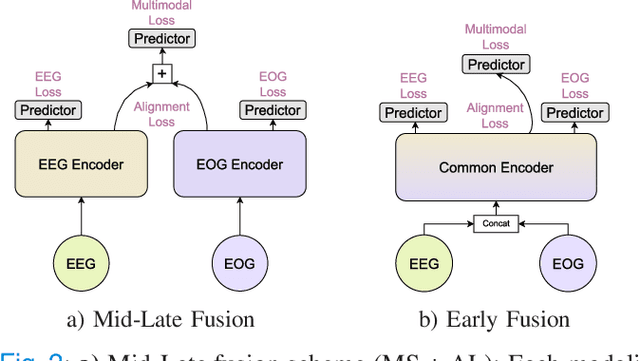
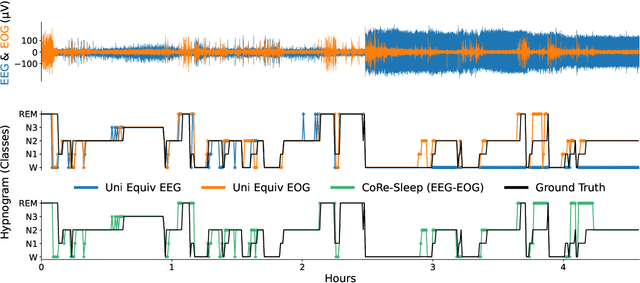
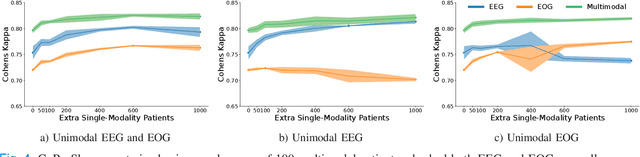
Abstract:Sleep abnormalities can have severe health consequences. Automated sleep staging, i.e. labelling the sequence of sleep stages from the patient's physiological recordings, could simplify the diagnostic process. Previous work on automated sleep staging has achieved great results, mainly relying on the EEG signal. However, often multiple sources of information are available beyond EEG. This can be particularly beneficial when the EEG recordings are noisy or even missing completely. In this paper, we propose CoRe-Sleep, a Coordinated Representation multimodal fusion network that is particularly focused on improving the robustness of signal analysis on imperfect data. We demonstrate how appropriately handling multimodal information can be the key to achieving such robustness. CoRe-Sleep tolerates noisy or missing modalities segments, allowing training on incomplete data. Additionally, it shows state-of-the-art performance when testing on both multimodal and unimodal data using a single model on SHHS-1, the largest publicly available study that includes sleep stage labels. The results indicate that training the model on multimodal data does positively influence performance when tested on unimodal data. This work aims at bridging the gap between automated analysis tools and their clinical utility.
Automated Movement Detection with Dirichlet Process Mixture Models and Electromyography
Feb 15, 2023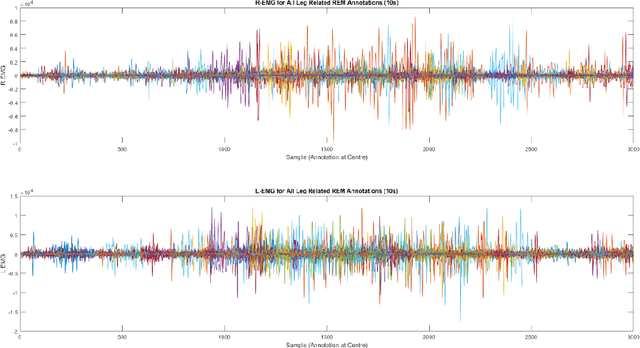
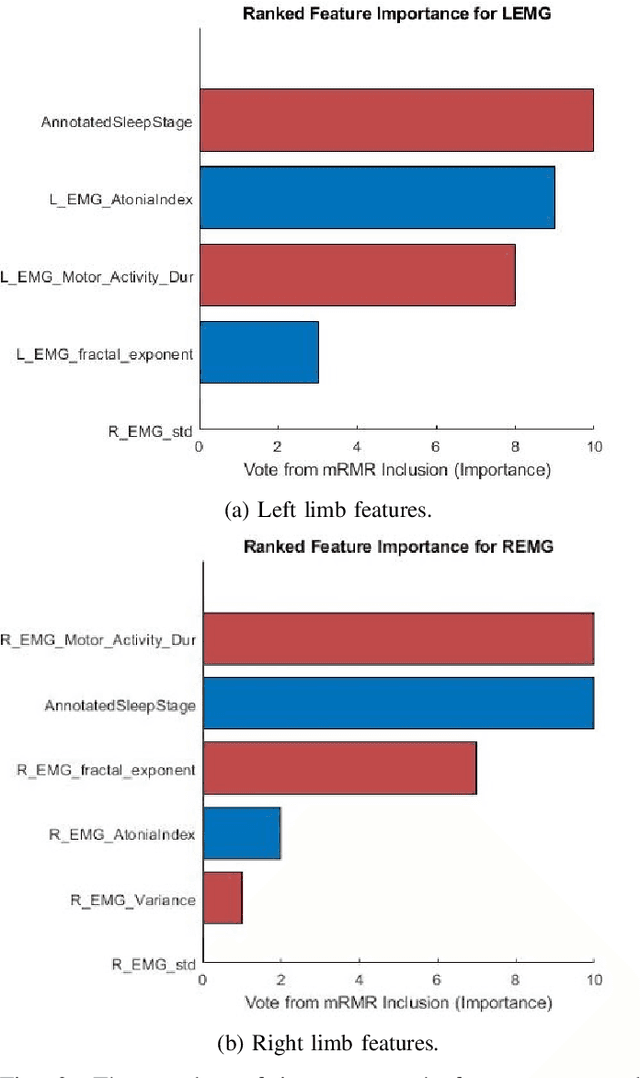
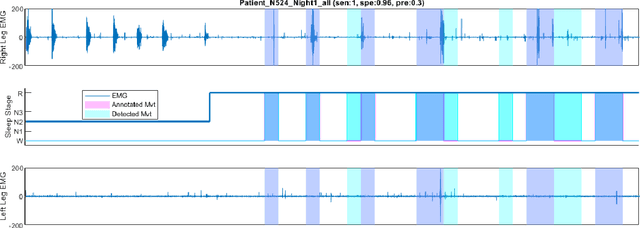

Abstract:Numerous sleep disorders are characterised by movement during sleep, these include rapid-eye movement sleep behaviour disorder (RBD) and periodic limb movement disorder. The process of diagnosing movement related sleep disorders requires laborious and time-consuming visual analysis of sleep recordings. This process involves sleep clinicians visually inspecting electromyogram (EMG) signals to identify abnormal movements. The distribution of characteristics that represent movement can be diverse and varied, ranging from brief moments of tensing to violent outbursts. This study proposes a framework for automated limb-movement detection by fusing data from two EMG sensors (from the left and right limb) through a Dirichlet process mixture model. Several features are extracted from 10 second mini-epochs, where each mini-epoch has been classified as 'leg-movement' or 'no leg-movement' based on annotations of movement from sleep clinicians. The distributions of the features from each category can be estimated accurately using Gaussian mixture models with the Dirichlet process as a prior. The available dataset includes 36 participants that have all been diagnosed with RBD. The performance of this framework was evaluated by a 10-fold cross validation scheme (participant independent). The study was compared to a random forest model and outperformed it with a mean accuracy, sensitivity, and specificity of 94\%, 48\%, and 95\%, respectively. These results demonstrate the ability of this framework to automate the detection of limb movement for the potential application of assisting clinical diagnosis and decision-making.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge