Christine Lo
Nuffield Department of Clinical Neurosciences, Oxford Parkinson's Disease Centre
Automated Movement Detection with Dirichlet Process Mixture Models and Electromyography
Feb 15, 2023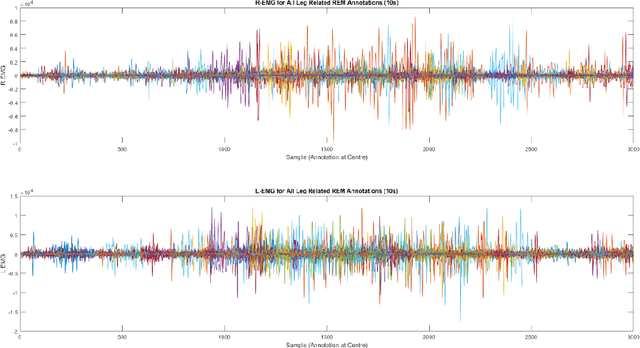
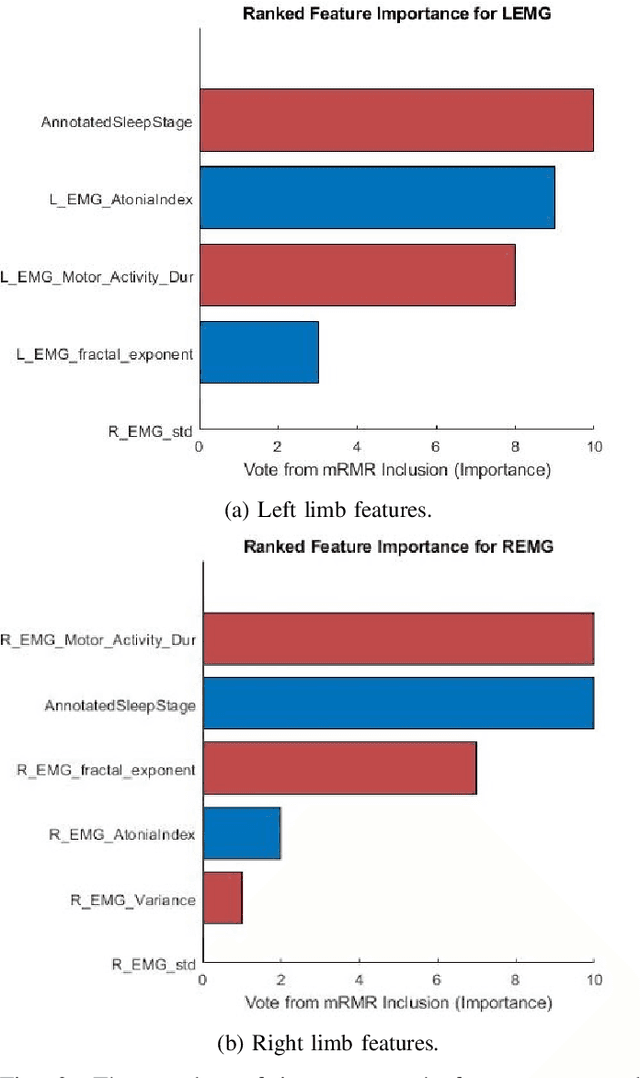
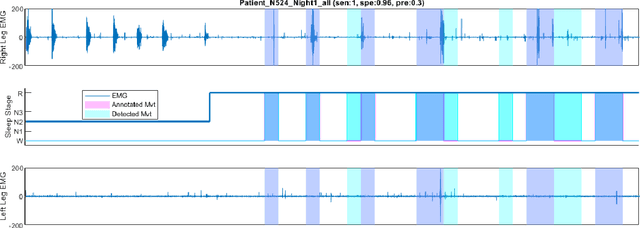

Abstract:Numerous sleep disorders are characterised by movement during sleep, these include rapid-eye movement sleep behaviour disorder (RBD) and periodic limb movement disorder. The process of diagnosing movement related sleep disorders requires laborious and time-consuming visual analysis of sleep recordings. This process involves sleep clinicians visually inspecting electromyogram (EMG) signals to identify abnormal movements. The distribution of characteristics that represent movement can be diverse and varied, ranging from brief moments of tensing to violent outbursts. This study proposes a framework for automated limb-movement detection by fusing data from two EMG sensors (from the left and right limb) through a Dirichlet process mixture model. Several features are extracted from 10 second mini-epochs, where each mini-epoch has been classified as 'leg-movement' or 'no leg-movement' based on annotations of movement from sleep clinicians. The distributions of the features from each category can be estimated accurately using Gaussian mixture models with the Dirichlet process as a prior. The available dataset includes 36 participants that have all been diagnosed with RBD. The performance of this framework was evaluated by a 10-fold cross validation scheme (participant independent). The study was compared to a random forest model and outperformed it with a mean accuracy, sensitivity, and specificity of 94\%, 48\%, and 95\%, respectively. These results demonstrate the ability of this framework to automate the detection of limb movement for the potential application of assisting clinical diagnosis and decision-making.
Detection of REM Sleep Behaviour Disorder by Automated Polysomnography Analysis
Nov 12, 2018
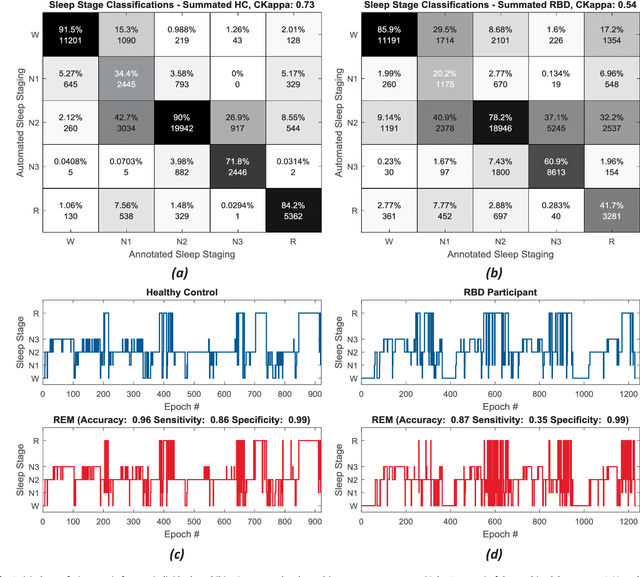
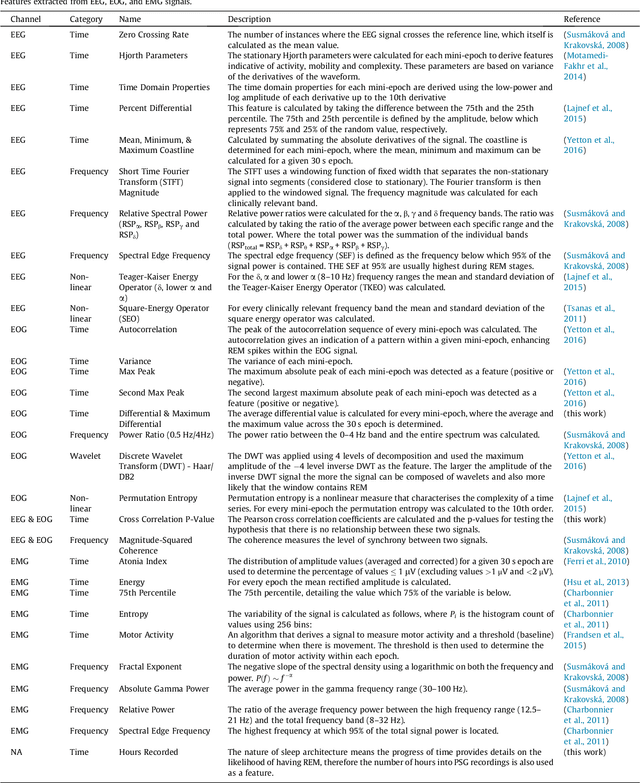
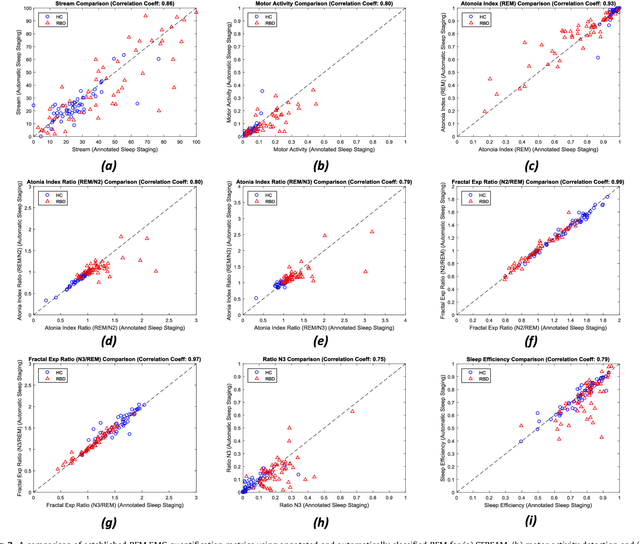
Abstract:Evidence suggests Rapid-Eye-Movement (REM) Sleep Behaviour Disorder (RBD) is an early predictor of Parkinson's disease. This study proposes a fully-automated framework for RBD detection consisting of automated sleep staging followed by RBD identification. Analysis was assessed using a limited polysomnography montage from 53 participants with RBD and 53 age-matched healthy controls. Sleep stage classification was achieved using a Random Forest (RF) classifier and 156 features extracted from electroencephalogram (EEG), electrooculogram (EOG) and electromyogram (EMG) channels. For RBD detection, a RF classifier was trained combining established techniques to quantify muscle atonia with additional features that incorporate sleep architecture and the EMG fractal exponent. Automated multi-state sleep staging achieved a 0.62 Cohen's Kappa score. RBD detection accuracy improved by 10% to 96% (compared to individual established metrics) when using manually annotated sleep staging. Accuracy remained high (92%) when using automated sleep staging. This study outperforms established metrics and demonstrates that incorporating sleep architecture and sleep stage transitions can benefit RBD detection. This study also achieved automated sleep staging with a level of accuracy comparable to manual annotation. This study validates a tractable, fully-automated, and sensitive pipeline for RBD identification that could be translated to wearable take-home technology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge