Danny Eytan
Aiming for Relevance
Mar 27, 2024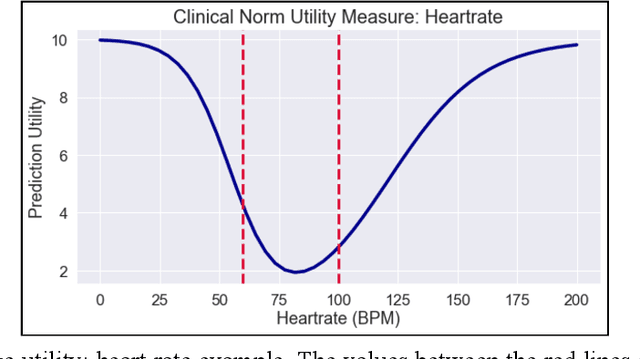
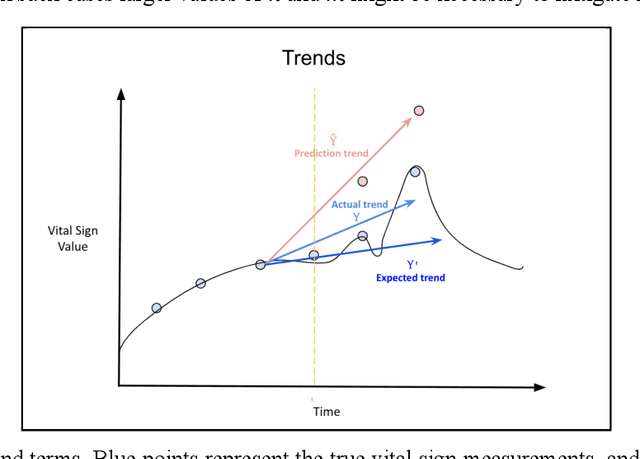
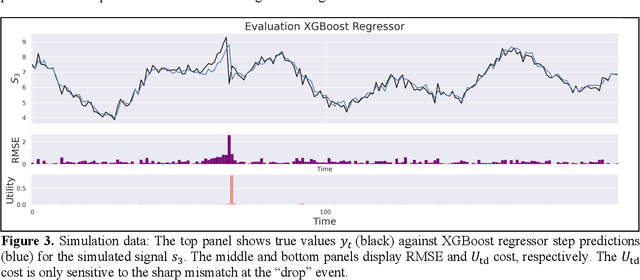
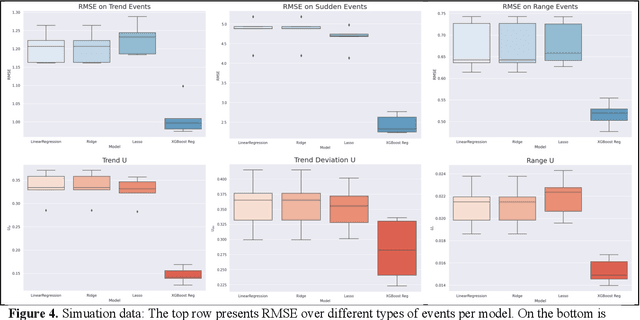
Abstract:Vital signs are crucial in intensive care units (ICUs). They are used to track the patient's state and to identify clinically significant changes. Predicting vital sign trajectories is valuable for early detection of adverse events. However, conventional machine learning metrics like RMSE often fail to capture the true clinical relevance of such predictions. We introduce novel vital sign prediction performance metrics that align with clinical contexts, focusing on deviations from clinical norms, overall trends, and trend deviations. These metrics are derived from empirical utility curves obtained in a previous study through interviews with ICU clinicians. We validate the metrics' usefulness using simulated and real clinical datasets (MIMIC and eICU). Furthermore, we employ these metrics as loss functions for neural networks, resulting in models that excel in predicting clinically significant events. This research paves the way for clinically relevant machine learning model evaluation and optimization, promising to improve ICU patient care. 10 pages, 9 figures.
Individualized Dosing Dynamics via Neural Eigen Decomposition
Jun 24, 2023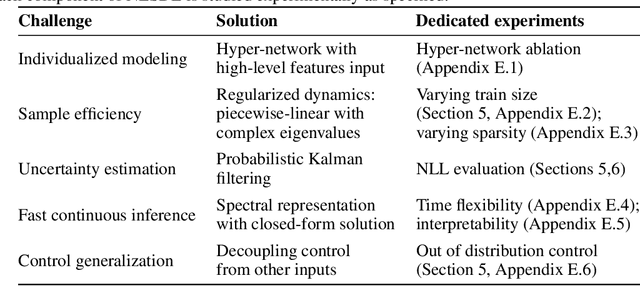



Abstract:Dosing models often use differential equations to model biological dynamics. Neural differential equations in particular can learn to predict the derivative of a process, which permits predictions at irregular points of time. However, this temporal flexibility often comes with a high sensitivity to noise, whereas medical problems often present high noise and limited data. Moreover, medical dosing models must generalize reliably over individual patients and changing treatment policies. To address these challenges, we introduce the Neural Eigen Stochastic Differential Equation algorithm (NESDE). NESDE provides individualized modeling (using a hypernetwork over patient-level parameters); generalization to new treatment policies (using decoupled control); tunable expressiveness according to the noise level (using piecewise linearity); and fast, continuous, closed-form prediction (using spectral representation). We demonstrate the robustness of NESDE in both synthetic and real medical problems, and use the learned dynamics to publish simulated medical gym environments.
Machine Learning to Support Triage of Children at Risk for Epileptic Seizures in the Pediatric Intensive Care Unit
May 11, 2022



Abstract:Objective: Epileptic seizures are relatively common in critically-ill children admitted to the pediatric intensive care unit (PICU) and thus serve as an important target for identification and treatment. Most of these seizures have no discernible clinical manifestation but still have a significant impact on morbidity and mortality. Children that are deemed at risk for seizures within the PICU are monitored using continuous-electroencephalogram (cEEG). cEEG monitoring cost is considerable and as the number of available machines is always limited, clinicians need to resort to triaging patients according to perceived risk in order to allocate resources. This research aims to develop a computer aided tool to improve seizures risk assessment in critically-ill children, using an ubiquitously recorded signal in the PICU, namely the electrocardiogram (ECG). Approach: A novel data-driven model was developed at a patient-level approach, based on features extracted from the first hour of ECG recording and the clinical data of the patient. Main results: The most predictive features were the age of the patient, the brain injury as coma etiology and the QRS area. For patients without any prior clinical data, using one hour of ECG recording, the classification performance of the random forest classifier reached an area under the receiver operating characteristic curve (AUROC) score of 0.84. When combining ECG features with the patients clinical history, the AUROC reached 0.87. Significance: Taking a real clinical scenario, we estimated that our clinical decision support triage tool can improve the positive predictive value by more than 59% over the clinical standard.
About Explicit Variance Minimization: Training Neural Networks for Medical Imaging With Limited Data Annotations
Jun 03, 2021



Abstract:Self-supervised learning methods for computer vision have demonstrated the effectiveness of pre-training feature representations, resulting in well-generalizing Deep Neural Networks, even if the annotated data are limited. However, representation learning techniques require a significant amount of time for model training, with most of it time spent on precise hyper-parameter optimization and selection of augmentation techniques. We hypothesized that if the annotated dataset has enough morphological diversity to capture the general population's as is common in medical imaging, for example, due to conserved similarities of tissue mythologies, the variance error of the trained model is the prevalent component of the Bias-Variance Trade-off. We propose the Variance Aware Training (VAT) method that exploits this property by introducing the variance error into the model loss function, i.e., enabling minimizing the variance explicitly. Additionally, we provide the theoretical formulation and proof of the proposed method to aid in interpreting the approach. Our method requires selecting only one hyper-parameter and was able to match or improve the state-of-the-art performance of self-supervised methods while achieving an order of magnitude reduction in the GPU training time. We validated VAT on three medical imaging datasets from diverse domains and various learning objectives. These included a Magnetic Resonance Imaging (MRI) dataset for the heart semantic segmentation (MICCAI 2017 ACDC challenge), fundus photography dataset for ordinary regression of diabetic retinopathy progression (Kaggle 2019 APTOS Blindness Detection challenge), and classification of histopathologic scans of lymph node sections (PatchCamelyon dataset).
Unsupervised Representation Learning for Time Series with Temporal Neighborhood Coding
Jun 01, 2021



Abstract:Time series are often complex and rich in information but sparsely labeled and therefore challenging to model. In this paper, we propose a self-supervised framework for learning generalizable representations for non-stationary time series. Our approach, called Temporal Neighborhood Coding (TNC), takes advantage of the local smoothness of a signal's generative process to define neighborhoods in time with stationary properties. Using a debiased contrastive objective, our framework learns time series representations by ensuring that in the encoding space, the distribution of signals from within a neighborhood is distinguishable from the distribution of non-neighboring signals. Our motivation stems from the medical field, where the ability to model the dynamic nature of time series data is especially valuable for identifying, tracking, and predicting the underlying patients' latent states in settings where labeling data is practically impossible. We compare our method to recently developed unsupervised representation learning approaches and demonstrate superior performance on clustering and classification tasks for multiple datasets.
Using Deep Networks for Scientific Discovery in Physiological Signals
Aug 25, 2020



Abstract:Deep neural networks (DNN) have shown remarkable success in the classification of physiological signals. In this study we propose a method for examining to what extent does a DNN's performance rely on rediscovering existing features of the signals, as opposed to discovering genuinely new features. Moreover, we offer a novel method of "removing" a hand-engineered feature from the network's hypothesis space, thus forcing it to try and learn representations which are different from known ones, as a method of scientific exploration. We then build on existing work in the field of interpretability, specifically class activation maps, to try and infer what new features the network has learned. We demonstrate this approach using ECG and EEG signals. With respect to ECG signals we show that for the specific task of classifying atrial fibrillation, DNNs are likely rediscovering known features. We also show how our method could be used to discover new features, by selectively removing some ECG features and "rediscovering" them. We further examine how could our method be used as a tool for examining scientific hypotheses. We simulate this scenario by looking into the importance of eye movements in classifying sleep from EEG. We show that our tool can successfully focus a researcher's attention by bringing to light patterns in the data that would be hidden otherwise.
Generative ODE Modeling with Known Unknowns
Mar 24, 2020



Abstract:In several crucial applications, domain knowledge is encoded by a system of ordinary differential equations (ODE). A motivating example is intensive care unit patients: The dynamics of some vital physiological variables such as heart rate, blood pressure and arterial compliance can be approximately described by a known system of ODEs. Typically, some of the ODE variables are directly observed while some are unobserved, and in addition many other variables are observed but not modeled by the ODE, for example body temperature. Importantly, the unobserved ODE variables are ``known-unknowns'': We know they exist and their functional dynamics, but cannot measure them directly, nor do we know the function tying them to all observed measurements. Estimating these known-unknowns is often highly valuable to physicians. Under this scenario we wish to: (i) learn the static parameters of the ODE generating each observed time-series (ii) infer the dynamic sequence of all ODE variables including the known-unknowns, and (iii) extrapolate the future of the ODE variables and the observations of the time-series. We address this task with a variational autoencoder incorporating the known ODE function, called GOKU-net for Generative ODE modeling with Known Unknowns. We test our method on videos of pendulums with unknown length, and a model of the cardiovascular system.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge