Seyed Mostafa Kia
PROMISSING: Pruning Missing Values in Neural Networks
Jun 03, 2022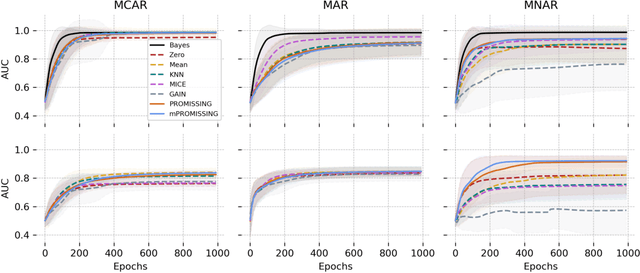
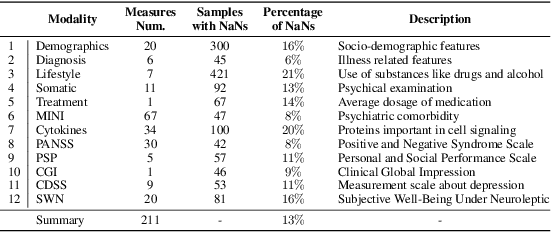
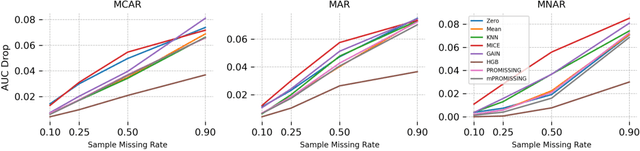
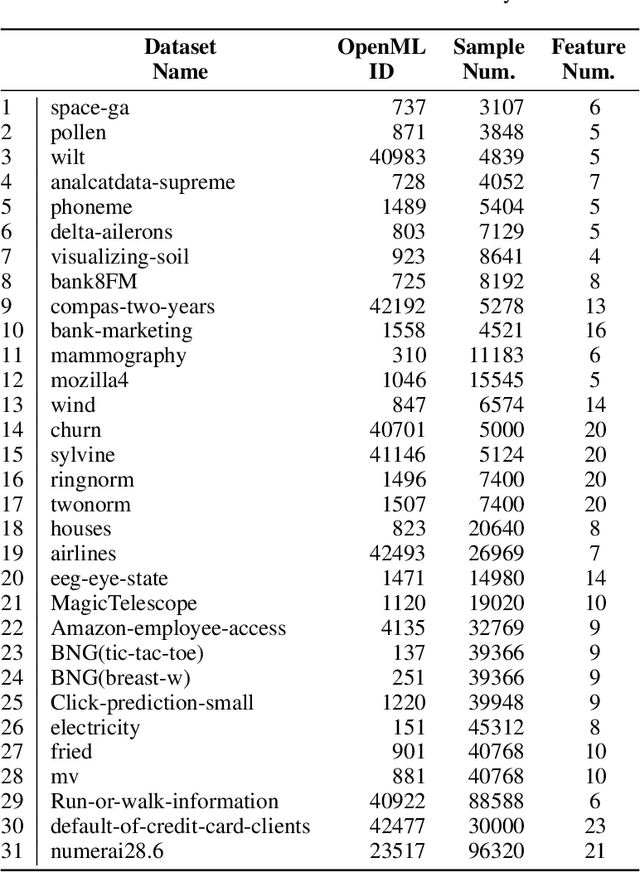
Abstract:While data are the primary fuel for machine learning models, they often suffer from missing values, especially when collected in real-world scenarios. However, many off-the-shelf machine learning models, including artificial neural network models, are unable to handle these missing values directly. Therefore, extra data preprocessing and curation steps, such as data imputation, are inevitable before learning and prediction processes. In this study, we propose a simple and intuitive yet effective method for pruning missing values (PROMISSING) during learning and inference steps in neural networks. In this method, there is no need to remove or impute the missing values; instead, the missing values are treated as a new source of information (representing what we do not know). Our experiments on simulated data, several classification and regression benchmarks, and a multi-modal clinical dataset show that PROMISSING results in similar prediction performance compared to various imputation techniques. In addition, our experiments show models trained using PROMISSING techniques are becoming less decisive in their predictions when facing incomplete samples with many unknowns. This finding hopefully advances machine learning models from being pure predicting machines to more realistic thinkers that can also say "I do not know" when facing incomplete sources of information.
Hierarchical Bayesian Regression for Multi-Site Normative Modeling of Neuroimaging Data
May 25, 2020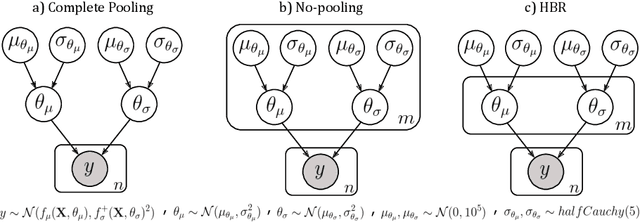

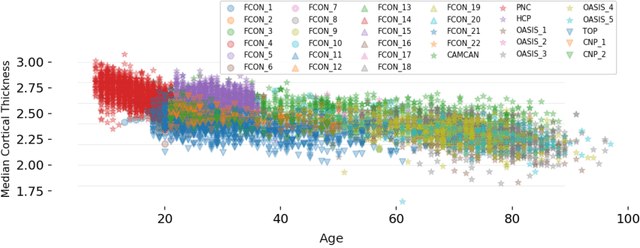
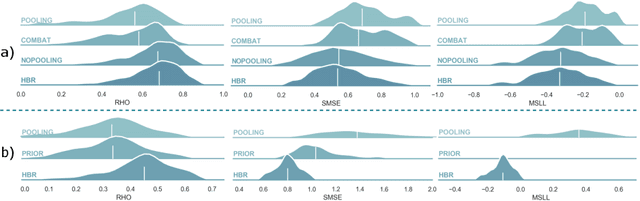
Abstract:Clinical neuroimaging has recently witnessed explosive growth in data availability which brings studying heterogeneity in clinical cohorts to the spotlight. Normative modeling is an emerging statistical tool for achieving this objective. However, its application remains technically challenging due to difficulties in properly dealing with nuisance variation, for example due to variability in image acquisition devices. Here, in a fully probabilistic framework, we propose an application of hierarchical Bayesian regression (HBR) for multi-site normative modeling. Our experimental results confirm the superiority of HBR in deriving more accurate normative ranges on large multi-site neuroimaging data compared to widely used methods. This provides the possibility i) to learn the normative range of structural and functional brain measures on large multi-site data; ii) to recalibrate and reuse the learned model on local small data; therefore, HBR closes the technical loop for applying normative modeling as a medical tool for the diagnosis and prognosis of mental disorders.
Neural Processes Mixed-Effect Models for Deep Normative Modeling of Clinical Neuroimaging Data
Dec 12, 2018



Abstract:Normative modeling has recently been introduced as a promising approach for modeling variation of neuroimaging measures across individuals in order to derive biomarkers of psychiatric disorders. Current implementations rely on Gaussian process regression, which provides coherent estimates of uncertainty needed for the method but also suffers from drawbacks including poor scaling to large datasets and a reliance on fixed parametric kernels. In this paper, we propose a deep normative modeling framework based on neural processes (NPs) to solve these problems. To achieve this, we define a stochastic process formulation for mixed-effect models and show how NPs can be adopted for spatially structured mixed-effect modeling of neuroimaging data. This enables us to learn optimal feature representations and covariance structure for the random-effect and noise via global latent variables. In this scheme, predictive uncertainty can be approximated by sampling from the distribution of these global latent variables. On a publicly available clinical fMRI dataset, we compare the novelty detection performance of multivariate normative models estimated by the proposed NP approach to a baseline multi-task Gaussian process regression approach and show substantial improvements for certain diagnostic problems.
Scalable Multi-Task Gaussian Process Tensor Regression for Normative Modeling of Structured Variation in Neuroimaging Data
Jul 31, 2018



Abstract:Most brain disorders are very heterogeneous in terms of their underlying biology and developing analysis methods to model such heterogeneity is a major challenge. A promising approach is to use probabilistic regression methods to estimate normative models of brain function using (f)MRI data then use these to map variation across individuals in clinical populations (e.g., via anomaly detection). To fully capture individual differences, it is crucial to statistically model the patterns of correlation across different brain regions and individuals. However, this is very challenging for neuroimaging data because of high-dimensionality and highly structured patterns of correlation across multiple axes. Here, we propose a general and flexible multi-task learning framework to address this problem. Our model uses a tensor-variate Gaussian process in a Bayesian mixed-effects model and makes use of Kronecker algebra and a low-rank approximation to scale efficiently to multi-way neuroimaging data at the whole brain level. On a publicly available clinical fMRI dataset, we show that our computationally affordable approach substantially improves detection sensitivity over both a mass-univariate normative model and a classifier that --unlike our approach-- has full access to the clinical labels.
Normative Modeling of Neuroimaging Data using Scalable Multi-Task Gaussian Processes
Jun 05, 2018

Abstract:Normative modeling has recently been proposed as an alternative for the case-control approach in modeling heterogeneity within clinical cohorts. Normative modeling is based on single-output Gaussian process regression that provides coherent estimates of uncertainty required by the method but does not consider spatial covariance structure. Here, we introduce a scalable multi-task Gaussian process regression (S-MTGPR) approach to address this problem. To this end, we exploit a combination of a low-rank approximation of the spatial covariance matrix with algebraic properties of Kronecker product in order to reduce the computational complexity of Gaussian process regression in high-dimensional output spaces. On a public fMRI dataset, we show that S-MTGPR: 1) leads to substantial computational improvements that allow us to estimate normative models for high-dimensional fMRI data whilst accounting for spatial structure in data; 2) by modeling both spatial and across-sample variances, it provides higher sensitivity in novelty detection scenarios.
Deep Learning for Automatic Stereotypical Motor Movement Detection using Wearable Sensors in Autism Spectrum Disorders
Sep 14, 2017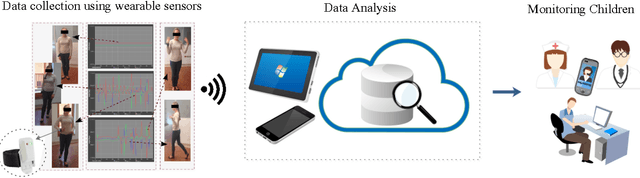
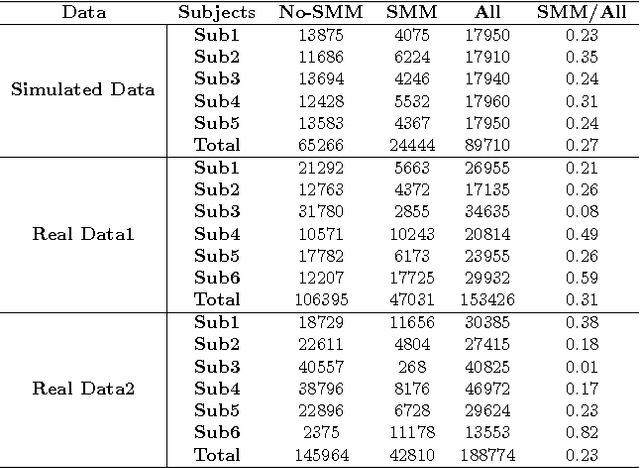
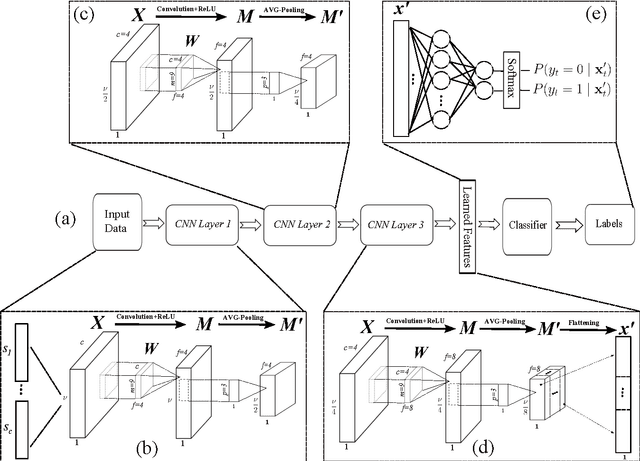
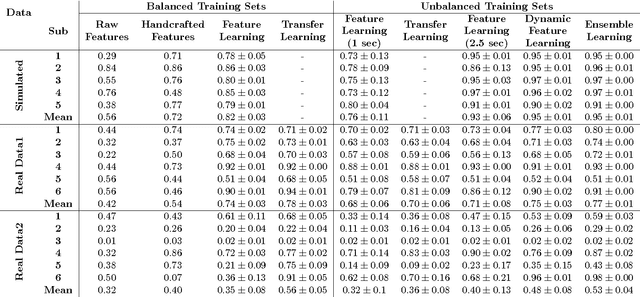
Abstract:Autism Spectrum Disorders are associated with atypical movements, of which stereotypical motor movements (SMMs) interfere with learning and social interaction. The automatic SMM detection using inertial measurement units (IMU) remains complex due to the strong intra and inter-subject variability, especially when handcrafted features are extracted from the signal. We propose a new application of the deep learning to facilitate automatic SMM detection using multi-axis IMUs. We use a convolutional neural network (CNN) to learn a discriminative feature space from raw data. We show how the CNN can be used for parameter transfer learning to enhance the detection rate on longitudinal data. We also combine the long short-term memory (LSTM) with CNN to model the temporal patterns in a sequence of multi-axis signals. Further, we employ ensemble learning to combine multiple LSTM learners into a more robust SMM detector. Our results show that: 1) feature learning outperforms handcrafted features; 2) parameter transfer learning is beneficial in longitudinal settings; 3) using LSTM to learn the temporal dynamic of signals enhances the detection rate especially for skewed training data; 4) an ensemble of LSTMs provides more accurate and stable detectors. These findings provide a significant step toward accurate SMM detection in real-time scenarios.
Interpretability in Linear Brain Decoding
Jun 17, 2016

Abstract:Improving the interpretability of brain decoding approaches is of primary interest in many neuroimaging studies. Despite extensive studies of this type, at present, there is no formal definition for interpretability of brain decoding models. As a consequence, there is no quantitative measure for evaluating the interpretability of different brain decoding methods. In this paper, we present a simple definition for interpretability of linear brain decoding models. Then, we propose to combine the interpretability and the performance of the brain decoding into a new multi-objective criterion for model selection. Our preliminary results on the toy data show that optimizing the hyper-parameters of the regularized linear classifier based on the proposed criterion results in more informative linear models. The presented definition provides the theoretical background for quantitative evaluation of interpretability in linear brain decoding.
Convolutional Neural Network for Stereotypical Motor Movement Detection in Autism
Jun 07, 2016
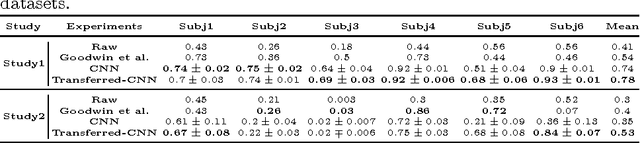

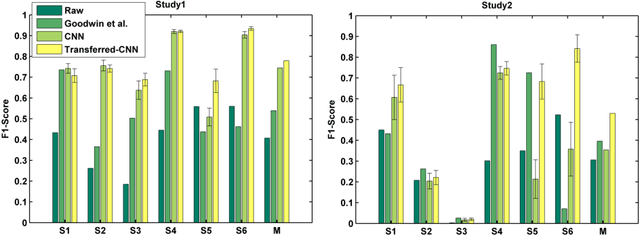
Abstract:Autism Spectrum Disorders (ASDs) are often associated with specific atypical postural or motor behaviors, of which Stereotypical Motor Movements (SMMs) have a specific visibility. While the identification and the quantification of SMM patterns remain complex, its automation would provide support to accurate tuning of the intervention in the therapy of autism. Therefore, it is essential to develop automatic SMM detection systems in a real world setting, taking care of strong inter-subject and intra-subject variability. Wireless accelerometer sensing technology can provide a valid infrastructure for real-time SMM detection, however such variability remains a problem also for machine learning methods, in particular whenever handcrafted features extracted from accelerometer signal are considered. Here, we propose to employ the deep learning paradigm in order to learn discriminating features from multi-sensor accelerometer signals. Our results provide preliminary evidence that feature learning and transfer learning embedded in the deep architecture achieve higher accurate SMM detectors in longitudinal scenarios.
Interpretability of Multivariate Brain Maps in Brain Decoding: Definition and Quantification
Mar 29, 2016



Abstract:Brain decoding is a popular multivariate approach for hypothesis testing in neuroimaging. It is well known that the brain maps derived from weights of linear classifiers are hard to interpret because of high correlations between predictors, low signal to noise ratios, and the high dimensionality of neuroimaging data. Therefore, improving the interpretability of brain decoding approaches is of primary interest in many neuroimaging studies. Despite extensive studies of this type, at present, there is no formal definition for interpretability of multivariate brain maps. As a consequence, there is no quantitative measure for evaluating the interpretability of different brain decoding methods. In this paper, first, we present a theoretical definition of interpretability in brain decoding; we show that the interpretability of multivariate brain maps can be decomposed into their reproducibility and representativeness. Second, as an application of the proposed theoretical definition, we formalize a heuristic method for approximating the interpretability of multivariate brain maps in a binary magnetoencephalography (MEG) decoding scenario. Third, we propose to combine the approximated interpretability and the performance of the brain decoding model into a new multi-objective criterion for model selection. Our results for the MEG data show that optimizing the hyper-parameters of the regularized linear classifier based on the proposed criterion results in more informative multivariate brain maps. More importantly, the presented definition provides the theoretical background for quantitative evaluation of interpretability, and hence, facilitates the development of more effective brain decoding algorithms in the future.
A Novel Scheme for Intelligent Recognition of Pornographic Images
Sep 29, 2014



Abstract:Harmful contents are rising in internet day by day and this motivates the essence of more research in fast and reliable obscene and immoral material filtering. Pornographic image recognition is an important component in each filtering system. In this paper, a new approach for detecting pornographic images is introduced. In this approach, two new features are suggested. These two features in combination with other simple traditional features provide decent difference between porn and non-porn images. In addition, we applied fuzzy integral based information fusion to combine MLP (Multi-Layer Perceptron) and NF (Neuro-Fuzzy) outputs. To test the proposed method, performance of system was evaluated over 18354 download images from internet. The attained precision was 93% in TP and 8% in FP on training dataset, and 87% and 5.5% on test dataset. Achieved results verify the performance of proposed system versus other related works.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge