Nicolo Micheletti
Generating Synthetic Free-text Medical Records with Low Re-identification Risk using Masked Language Modeling
Sep 17, 2024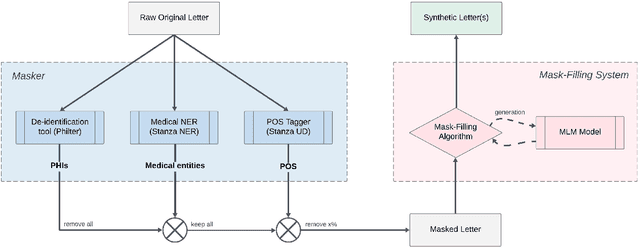
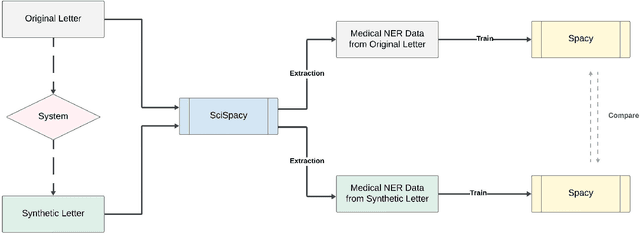

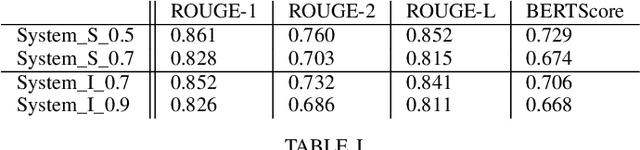
Abstract:In this paper, we present a system that generates synthetic free-text medical records, such as discharge summaries, admission notes and doctor correspondences, using Masked Language Modeling (MLM). Our system is designed to preserve the critical information of the records while introducing significant diversity and minimizing re-identification risk. The system incorporates a de-identification component that uses Philter to mask Protected Health Information (PHI), followed by a Medical Entity Recognition (NER) model to retain key medical information. We explore various masking ratios and mask-filling techniques to balance the trade-off between diversity and fidelity in the synthetic outputs without affecting overall readability. Our results demonstrate that the system can produce high-quality synthetic data with significant diversity while achieving a HIPAA-compliant PHI recall rate of 0.96 and a low re-identification risk of 0.035. Furthermore, downstream evaluations using a NER task reveal that the synthetic data can be effectively used to train models with performance comparable to those trained on real data. The flexibility of the system allows it to be adapted for specific use cases, making it a valuable tool for privacy-preserving data generation in medical research and healthcare applications.
BeeManc at the PLABA Track of TAC-2023: Investigating LLMs and Controllable Attributes for Improving Biomedical Text Readability
Aug 07, 2024



Abstract:In this system report, we describe the models and methods we used for our participation in the PLABA2023 task on biomedical abstract simplification, part of the TAC 2023 tracks. The system outputs we submitted come from the following three categories: 1) domain fine-tuned T5-like models including Biomedical-T5 and Lay-SciFive; 2) fine-tuned BARTLarge model with controllable attributes (via tokens) BART-w-CTs; 3) ChatGPTprompting. We also present the work we carried out for this task on BioGPT finetuning. In the official automatic evaluation using SARI scores, BeeManc ranks 2nd among all teams and our model LaySciFive ranks 3rd among all 13 evaluated systems. In the official human evaluation, our model BART-w-CTs ranks 2nd on Sentence-Simplicity (score 92.84), 3rd on Term-Simplicity (score 82.33) among all 7 evaluated systems; It also produced a high score 91.57 on Fluency in comparison to the highest score 93.53. In the second round of submissions, our team using ChatGPT-prompting ranks the 2nd in several categories including simplified term accuracy score 92.26 and completeness score 96.58, and a very similar score on faithfulness score 95.3 to re-evaluated PLABA-base-1 (95.73) via human evaluations. Our codes, fine-tuned models, prompts, and data splits from the system development stage will be available at https://github.com/ HECTA-UoM/PLABA-MU
Exploration of Masked and Causal Language Modelling for Text Generation
May 21, 2024Abstract:Large Language Models (LLMs) have revolutionised the field of Natural Language Processing (NLP) and have achieved state-of-the-art performance in practically every task in this field. However, the prevalent approach used in text generation, Causal Language Modelling (CLM), which generates text sequentially from left to right, inherently limits the freedom of the model, which does not decide when and where each token is generated. In contrast, Masked Language Modelling (MLM), primarily used for language understanding tasks, can generate tokens anywhere in the text and any order. This paper conducts an extensive comparison of MLM and CLM approaches for text generation tasks. To do so, we pre-train several language models of comparable sizes on three different datasets, namely 1) medical discharge summaries, 2) movie plot synopses, and 3) authorship verification datasets. To assess the quality of the generations, we first employ quantitative metrics and then perform a qualitative human evaluation to analyse coherence and grammatical correctness. In addition, we evaluate the usefulness of the generated texts by using them in three different downstream tasks: 1) Entity Recognition, 2) Text Classification, and 3) Authorship Verification. The results show that MLM consistently outperforms CLM in text generation across all datasets, with higher quantitative scores and better coherence in the generated text. The study also finds \textit{no strong correlation} between the quality of the generated text and the performance of the models in the downstream tasks. With this study, we show that MLM for text generation has great potential for future research and provides direction for future studies in this area.
Generating Medical Prescriptions with Conditional Transformer
Nov 18, 2023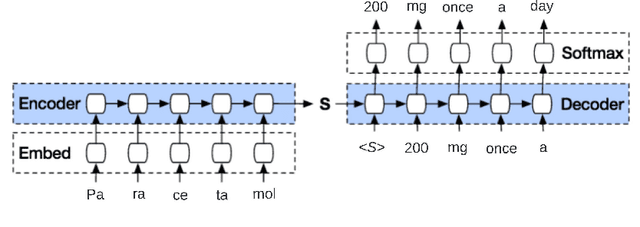
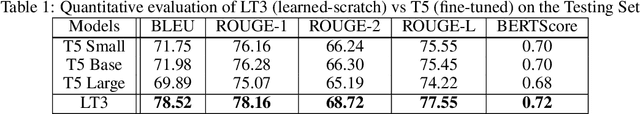
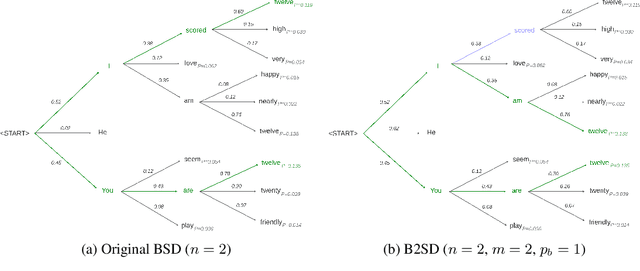

Abstract:Access to real-world medication prescriptions is essential for medical research and healthcare quality improvement. However, access to real medication prescriptions is often limited due to the sensitive nature of the information expressed. Additionally, manually labelling these instructions for training and fine-tuning Natural Language Processing (NLP) models can be tedious and expensive. We introduce a novel task-specific model architecture, Label-To-Text-Transformer (\textbf{LT3}), tailored to generate synthetic medication prescriptions based on provided labels, such as a vocabulary list of medications and their attributes. LT3 is trained on a set of around 2K lines of medication prescriptions extracted from the MIMIC-III database, allowing the model to produce valuable synthetic medication prescriptions. We evaluate LT3's performance by contrasting it with a state-of-the-art Pre-trained Language Model (PLM), T5, analysing the quality and diversity of generated texts. We deploy the generated synthetic data to train the SpacyNER model for the Named Entity Recognition (NER) task over the n2c2-2018 dataset. The experiments show that the model trained on synthetic data can achieve a 96-98\% F1 score at Label Recognition on Drug, Frequency, Route, Strength, and Form. LT3 codes and data will be shared at \url{https://github.com/HECTA-UoM/Label-To-Text-Transformer}
Large Language Models and Control Mechanisms Improve Text Readability of Biomedical Abstracts
Sep 22, 2023Abstract:Biomedical literature often uses complex language and inaccessible professional terminologies. That is why simplification plays an important role in improving public health literacy. Applying Natural Language Processing (NLP) models to automate such tasks allows for quick and direct accessibility for lay readers. In this work, we investigate the ability of state-of-the-art large language models (LLMs) on the task of biomedical abstract simplification, using the publicly available dataset for plain language adaptation of biomedical abstracts (\textbf{PLABA}). The methods applied include domain fine-tuning and prompt-based learning (PBL) on: 1) Encoder-decoder models (T5, SciFive, and BART), 2) Decoder-only GPT models (GPT-3.5 and GPT-4) from OpenAI and BioGPT, and 3) Control-token mechanisms on BART-based models. We used a range of automatic evaluation metrics, including BLEU, ROUGE, SARI, and BERTscore, and also conducted human evaluations. BART-Large with Control Token (BART-L-w-CT) mechanisms reported the highest SARI score of 46.54 and T5-base reported the highest BERTscore 72.62. In human evaluation, BART-L-w-CTs achieved a better simplicity score over T5-Base (2.9 vs. 2.2), while T5-Base achieved a better meaning preservation score over BART-L-w-CTs (3.1 vs. 2.6). We also categorised the system outputs with examples, hoping this will shed some light for future research on this task. Our code, fine-tuned models, and data splits are available at \url{https://github.com/HECTA-UoM/PLABA-MU}
Mitigating Health Data Poverty: Generative Approaches versus Resampling for Time-series Clinical Data
Oct 26, 2022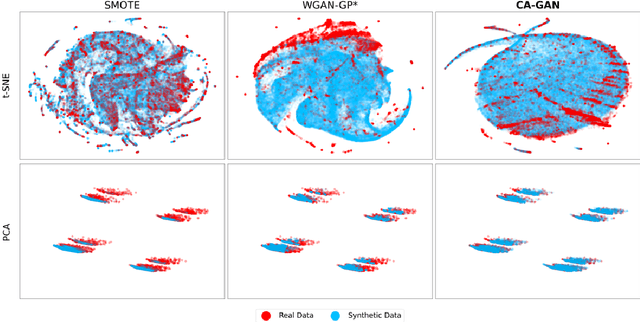
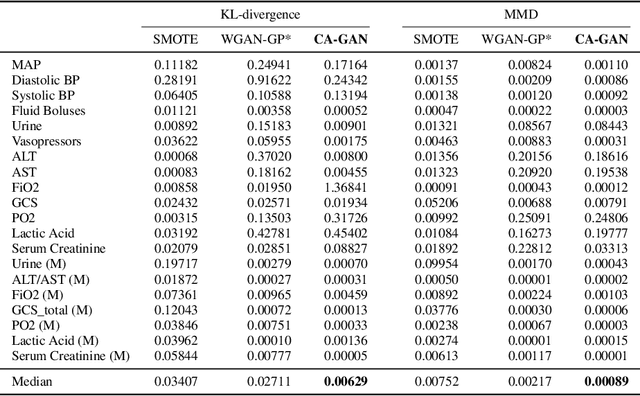
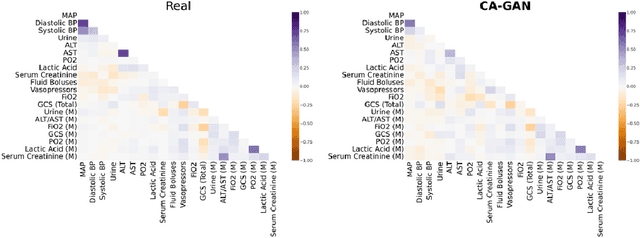
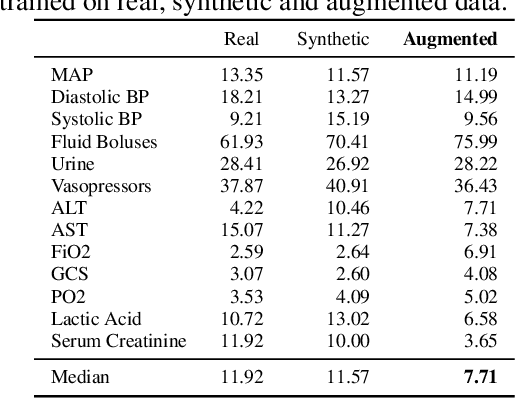
Abstract:Several approaches have been developed to mitigate algorithmic bias stemming from health data poverty, where minority groups are underrepresented in training datasets. Augmenting the minority class using resampling (such as SMOTE) is a widely used approach due to the simplicity of the algorithms. However, these algorithms decrease data variability and may introduce correlations between samples, giving rise to the use of generative approaches based on GAN. Generation of high-dimensional, time-series, authentic data that provides a wide distribution coverage of the real data, remains a challenging task for both resampling and GAN-based approaches. In this work we propose CA-GAN architecture that addresses some of the shortcomings of the current approaches, where we provide a detailed comparison with both SMOTE and WGAN-GP*, using a high-dimensional, time-series, real dataset of 3343 hypotensive Caucasian and Black patients. We show that our approach is better at both generating authentic data of the minority class and remaining within the original distribution of the real data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge