Warren Del-Pinto
SynBench: A Benchmark for Differentially Private Text Generation
Sep 18, 2025
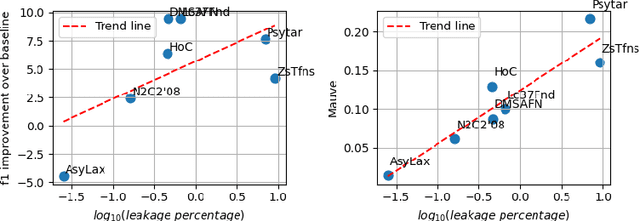
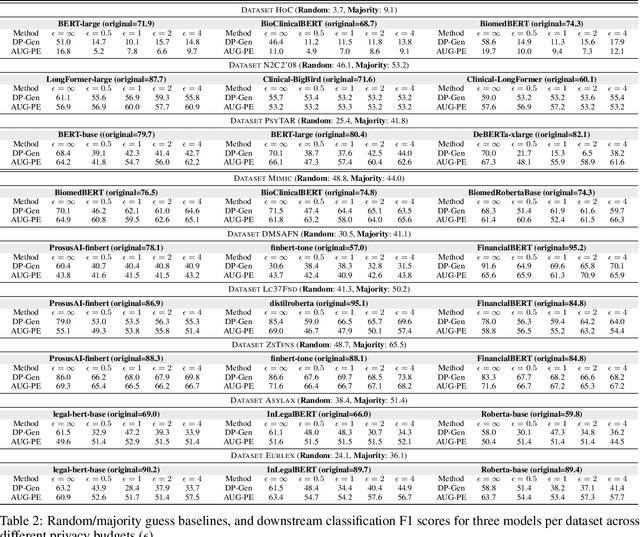
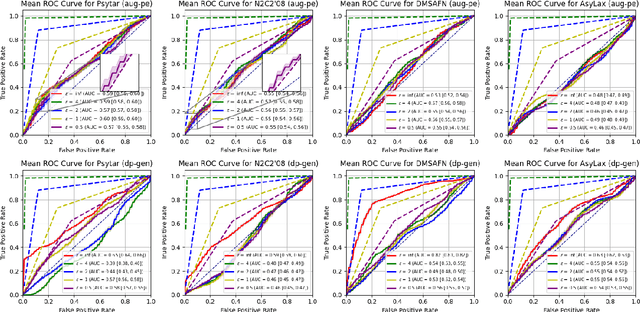
Abstract:Data-driven decision support in high-stakes domains like healthcare and finance faces significant barriers to data sharing due to regulatory, institutional, and privacy concerns. While recent generative AI models, such as large language models, have shown impressive performance in open-domain tasks, their adoption in sensitive environments remains limited by unpredictable behaviors and insufficient privacy-preserving datasets for benchmarking. Existing anonymization methods are often inadequate, especially for unstructured text, as redaction and masking can still allow re-identification. Differential Privacy (DP) offers a principled alternative, enabling the generation of synthetic data with formal privacy assurances. In this work, we address these challenges through three key contributions. First, we introduce a comprehensive evaluation framework with standardized utility and fidelity metrics, encompassing nine curated datasets that capture domain-specific complexities such as technical jargon, long-context dependencies, and specialized document structures. Second, we conduct a large-scale empirical study benchmarking state-of-the-art DP text generation methods and LLMs of varying sizes and different fine-tuning strategies, revealing that high-quality domain-specific synthetic data generation under DP constraints remains an unsolved challenge, with performance degrading as domain complexity increases. Third, we develop a membership inference attack (MIA) methodology tailored for synthetic text, providing first empirical evidence that the use of public datasets - potentially present in pre-training corpora - can invalidate claimed privacy guarantees. Our findings underscore the urgent need for rigorous privacy auditing and highlight persistent gaps between open-domain and specialist evaluations, informing responsible deployment of generative AI in privacy-sensitive, high-stakes settings.
DeIDClinic: A Multi-Layered Framework for De-identification of Clinical Free-text Data
Oct 02, 2024Abstract:De-identification is important in protecting patients' privacy for healthcare text analytics. The MASK framework is one of the best on the de-identification shared task organised by n2c2/i2b2 challenges. This work enhances the MASK framework by integrating ClinicalBERT, a deep learning model specifically fine-tuned on clinical texts, alongside traditional de-identification methods like dictionary lookup and rule-based approaches. The system effectively identifies and either redacts or replaces sensitive identifiable entities within clinical documents, while also allowing users to customise the masked documents according to their specific needs. The integration of ClinicalBERT significantly improves the performance of entity recognition, achieving 0.9732 F1-score, especially for common entities such as names, dates, and locations. A risk assessment feature has also been developed, which analyses the uniqueness of context within documents to classify them into risk levels, guiding further de-identification efforts. While the system demonstrates strong overall performance, this work highlights areas for future improvement, including handling more complex entity occurrences and enhancing the system's adaptability to different clinical settings.
Synthetic4Health: Generating Annotated Synthetic Clinical Letters
Sep 14, 2024Abstract:Since clinical letters contain sensitive information, clinical-related datasets can not be widely applied in model training, medical research, and teaching. This work aims to generate reliable, various, and de-identified synthetic clinical letters. To achieve this goal, we explored different pre-trained language models (PLMs) for masking and generating text. After that, we worked on Bio\_ClinicalBERT, a high-performing model, and experimented with different masking strategies. Both qualitative and quantitative methods were used for evaluation. Additionally, a downstream task, Named Entity Recognition (NER), was also implemented to assess the usability of these synthetic letters. The results indicate that 1) encoder-only models outperform encoder-decoder models. 2) Among encoder-only models, those trained on general corpora perform comparably to those trained on clinical data when clinical information is preserved. 3) Additionally, preserving clinical entities and document structure better aligns with our objectives than simply fine-tuning the model. 4) Furthermore, different masking strategies can impact the quality of synthetic clinical letters. Masking stopwords has a positive impact, while masking nouns or verbs has a negative effect. 5) For evaluation, BERTScore should be the primary quantitative evaluation metric, with other metrics serving as supplementary references. 6) Contextual information does not significantly impact the models' understanding, so the synthetic clinical letters have the potential to replace the original ones in downstream tasks.
Generating Medical Prescriptions with Conditional Transformer
Nov 18, 2023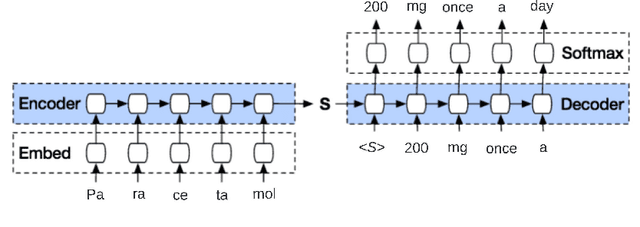
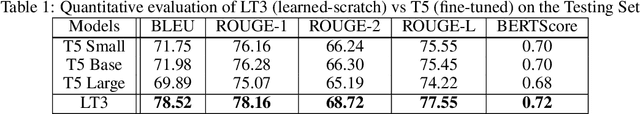

Abstract:Access to real-world medication prescriptions is essential for medical research and healthcare quality improvement. However, access to real medication prescriptions is often limited due to the sensitive nature of the information expressed. Additionally, manually labelling these instructions for training and fine-tuning Natural Language Processing (NLP) models can be tedious and expensive. We introduce a novel task-specific model architecture, Label-To-Text-Transformer (\textbf{LT3}), tailored to generate synthetic medication prescriptions based on provided labels, such as a vocabulary list of medications and their attributes. LT3 is trained on a set of around 2K lines of medication prescriptions extracted from the MIMIC-III database, allowing the model to produce valuable synthetic medication prescriptions. We evaluate LT3's performance by contrasting it with a state-of-the-art Pre-trained Language Model (PLM), T5, analysing the quality and diversity of generated texts. We deploy the generated synthetic data to train the SpacyNER model for the Named Entity Recognition (NER) task over the n2c2-2018 dataset. The experiments show that the model trained on synthetic data can achieve a 96-98\% F1 score at Label Recognition on Drug, Frequency, Route, Strength, and Form. LT3 codes and data will be shared at \url{https://github.com/HECTA-UoM/Label-To-Text-Transformer}
Exploring the Consistency, Quality and Challenges in Manual and Automated Coding of Free-text Diagnoses from Hospital Outpatient Letters
Nov 17, 2023
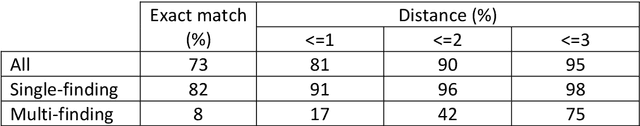
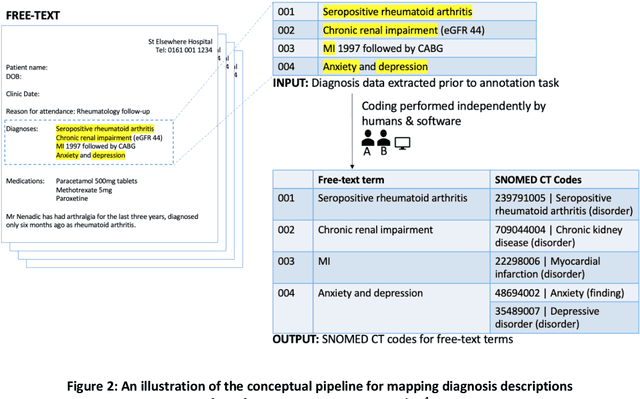
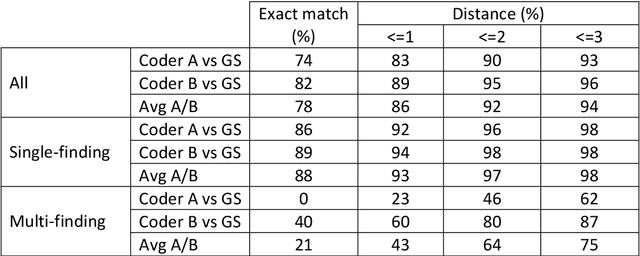
Abstract:Coding of unstructured clinical free-text to produce interoperable structured data is essential to improve direct care, support clinical communication and to enable clinical research.However, manual clinical coding is difficult and time consuming, which motivates the development and use of natural language processing for automated coding. This work evaluates the quality and consistency of both manual and automated clinical coding of diagnoses from hospital outpatient letters. Using 100 randomly selected letters, two human clinicians performed coding of diagnosis lists to SNOMED CT. Automated coding was also performed using IMO's Concept Tagger. A gold standard was constructed by a panel of clinicians from a subset of the annotated diagnoses. This was used to evaluate the quality and consistency of both manual and automated coding via (1) a distance-based metric, treating SNOMED CT as a graph, and (2) a qualitative metric agreed upon by the panel of clinicians. Correlation between the two metrics was also evaluated. Comparing human and computer-generated codes to the gold standard, the results indicate that humans slightly out-performed automated coding, while both performed notably better when there was only a single diagnosis contained in the free-text description. Automated coding was considered acceptable by the panel of clinicians in approximately 90% of cases.
Signature-Based Abduction for Expressive Description Logics -- Technical Report
Jul 08, 2020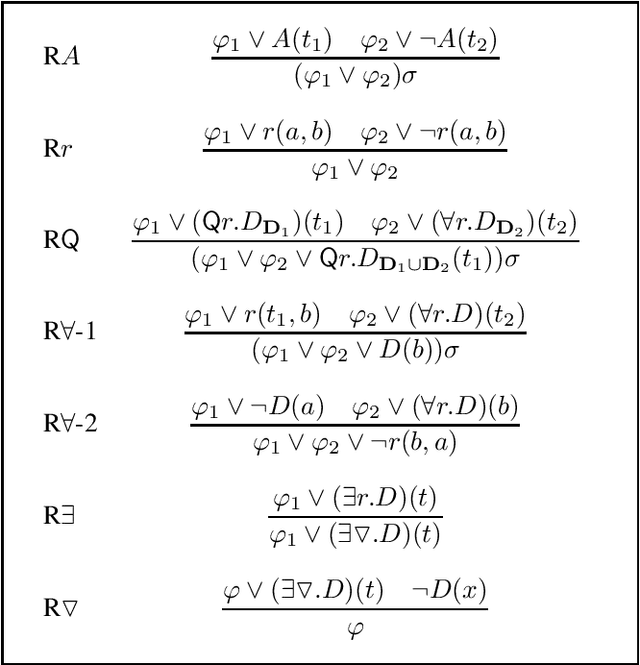
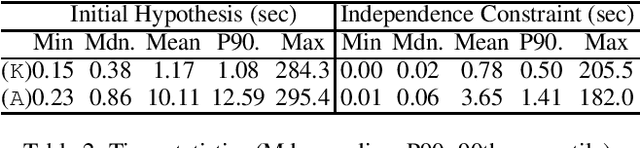
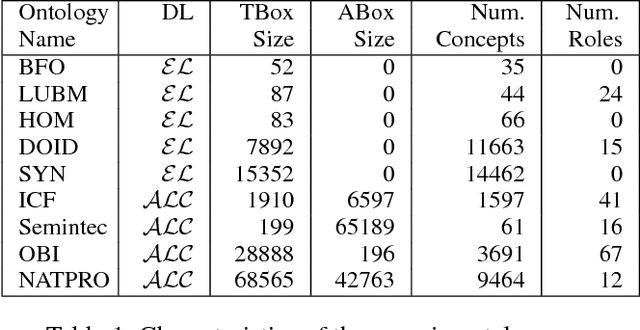
Abstract:Signature-based abduction aims at building hypotheses over a specified set of names, the signature, that explain an observation relative to some background knowledge. This type of abduction is useful for tasks such as diagnosis, where the vocabulary used for observed symptoms differs from the vocabulary expected to explain those symptoms. We present the first complete method solving signature-based abduction for observations expressed in the expressive description logic ALC, which can include TBox and ABox axioms, thereby solving the knowledge base abduction problem. The method is guaranteed to compute a finite and complete set of hypotheses, and is evaluated on a set of realistic knowledge bases.
ABox Abduction via Forgetting in ALC (Long Version)
Nov 13, 2018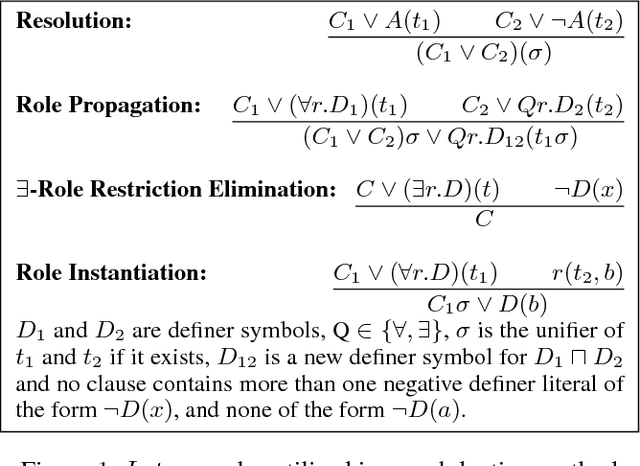
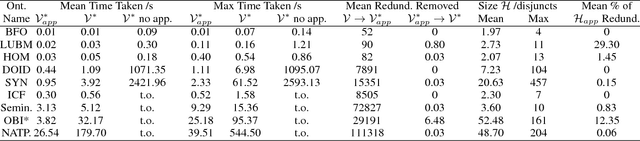
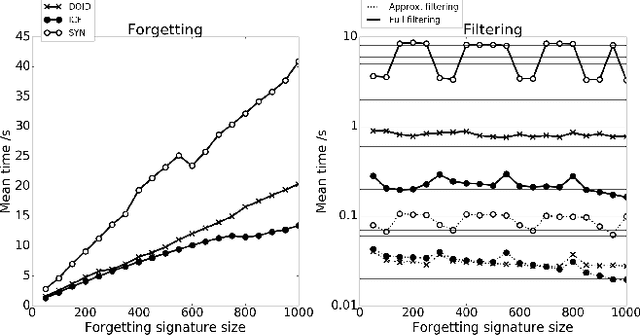
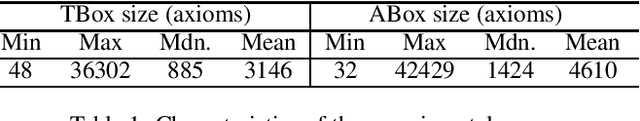
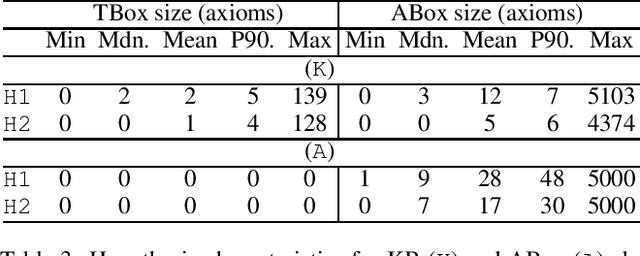
Abstract:Abductive reasoning generates explanatory hypotheses for new observations using prior knowledge. This paper investigates the use of forgetting, also known as uniform interpolation, to perform ABox abduction in description logic (ALC) ontologies. Non-abducibles are specified by a forgetting signature which can contain concept, but not role, symbols. The resulting hypotheses are semantically minimal and each consist of a set of disjuncts. These disjuncts are each independent explanations, and are not redundant with respect to the background ontology or the other disjuncts, representing a form of hypothesis space. The observations and hypotheses handled by the method can contain both atomic or complex ALC concepts, excluding role assertions, and are not restricted to Horn clauses. Two approaches to redundancy elimination are explored for practical use: full and approximate. Using a prototype implementation, experiments were performed over a corpus of real world ontologies to investigate the practicality of both approaches across several settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge