Niels Peek
Prediction of Survival Outcomes under Clinical Presence Shift: A Joint Neural Network Architecture
Aug 07, 2025Abstract:Electronic health records arise from the complex interaction between patients and the healthcare system. This observation process of interactions, referred to as clinical presence, often impacts observed outcomes. When using electronic health records to develop clinical prediction models, it is standard practice to overlook clinical presence, impacting performance and limiting the transportability of models when this interaction evolves. We propose a multi-task recurrent neural network that jointly models the inter-observation time and the missingness processes characterising this interaction in parallel to the survival outcome of interest. Our work formalises the concept of clinical presence shift when the prediction model is deployed in new settings (e.g. different hospitals, regions or countries), and we theoretically justify why the proposed joint modelling can improve transportability under changes in clinical presence. We demonstrate, in a real-world mortality prediction task in the MIMIC-III dataset, how the proposed strategy improves performance and transportability compared to state-of-the-art prediction models that do not incorporate the observation process. These results emphasise the importance of leveraging clinical presence to improve performance and create more transportable clinical prediction models.
Exploring the Consistency, Quality and Challenges in Manual and Automated Coding of Free-text Diagnoses from Hospital Outpatient Letters
Nov 17, 2023
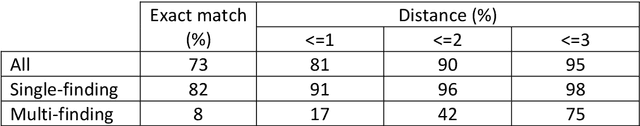
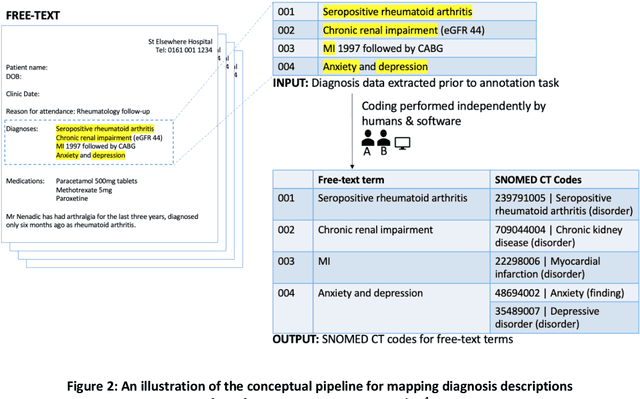
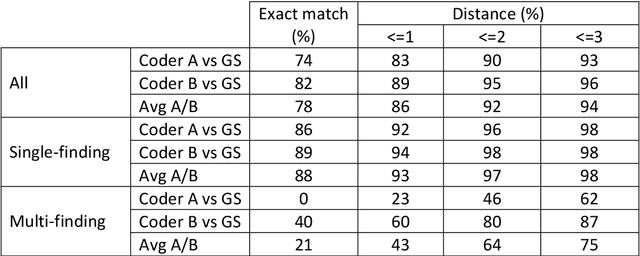
Abstract:Coding of unstructured clinical free-text to produce interoperable structured data is essential to improve direct care, support clinical communication and to enable clinical research.However, manual clinical coding is difficult and time consuming, which motivates the development and use of natural language processing for automated coding. This work evaluates the quality and consistency of both manual and automated clinical coding of diagnoses from hospital outpatient letters. Using 100 randomly selected letters, two human clinicians performed coding of diagnosis lists to SNOMED CT. Automated coding was also performed using IMO's Concept Tagger. A gold standard was constructed by a panel of clinicians from a subset of the annotated diagnoses. This was used to evaluate the quality and consistency of both manual and automated coding via (1) a distance-based metric, treating SNOMED CT as a graph, and (2) a qualitative metric agreed upon by the panel of clinicians. Correlation between the two metrics was also evaluated. Comparing human and computer-generated codes to the gold standard, the results indicate that humans slightly out-performed automated coding, while both performed notably better when there was only a single diagnosis contained in the free-text description. Automated coding was considered acceptable by the panel of clinicians in approximately 90% of cases.
DeepJoint: Robust Survival Modelling Under Clinical Presence Shift
May 26, 2022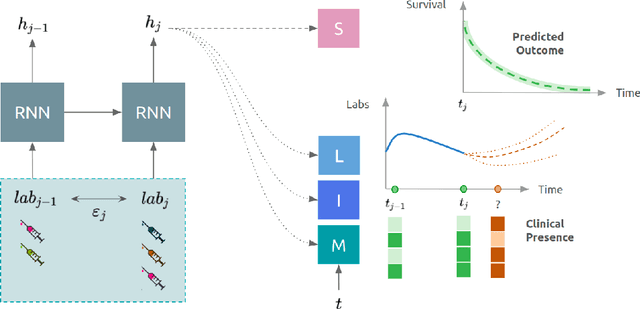
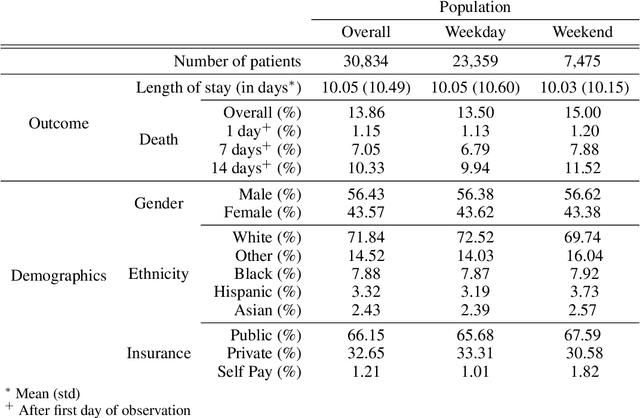
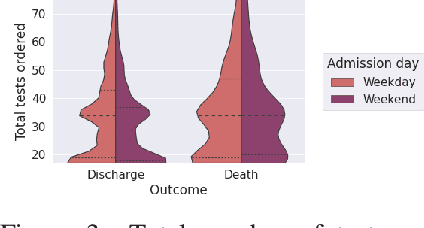
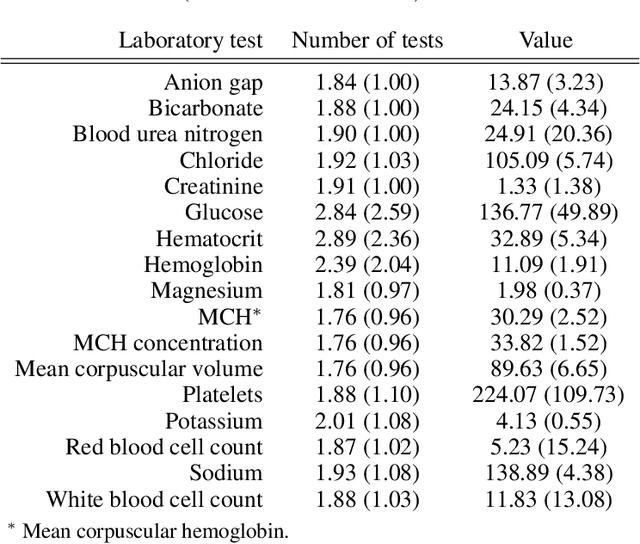
Abstract:Observational data in medicine arise as a result of the complex interaction between patients and the healthcare system. The sampling process is often highly irregular and itself constitutes an informative process. When using such data to develop prediction models, this phenomenon is often ignored, leading to sub-optimal performance and generalisability of models when practices evolve. We propose a multi-task recurrent neural network which models three clinical presence dimensions -- namely the longitudinal, the inter-observation and the missingness processes -- in parallel to the survival outcome. On a prediction task using MIMIC III laboratory tests, explicit modelling of these three processes showed improved performance in comparison to state-of-the-art predictive models (C-index at 1 day horizon: 0.878). More importantly, the proposed approach was more robust to change in the clinical presence setting, demonstrated by performance comparison between patients admitted on weekdays and weekends. This analysis demonstrates the importance of studying and leveraging clinical presence to improve performance and create more transportable clinical models.
EPICURE Ensemble Pretrained Models for Extracting Cancer Mutations from Literature
Jun 11, 2021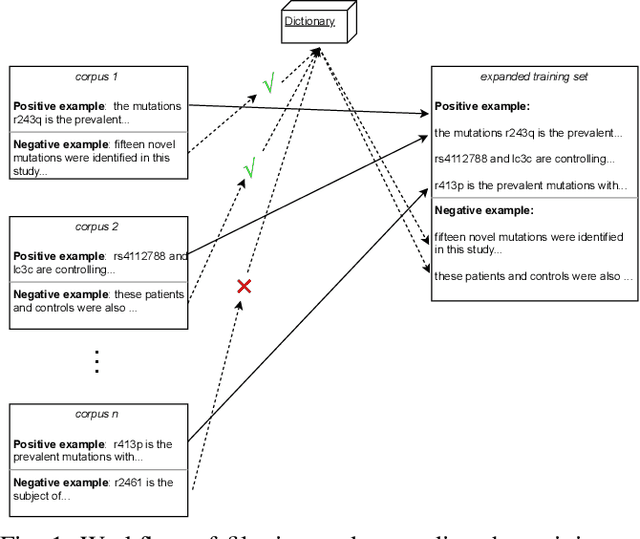
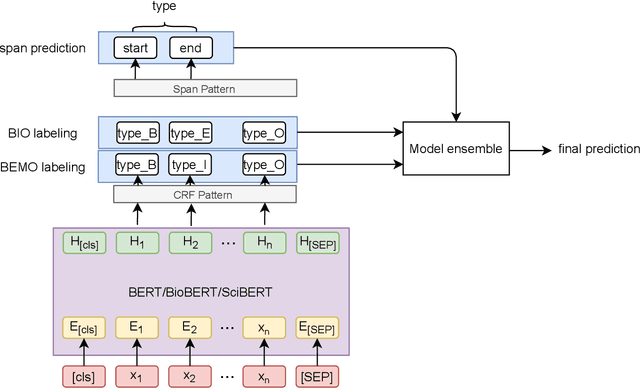

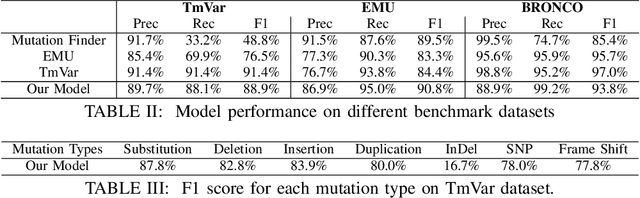
Abstract:To interpret the genetic profile present in a patient sample, it is necessary to know which mutations have important roles in the development of the corresponding cancer type. Named entity recognition is a core step in the text mining pipeline which facilitates mining valuable cancer information from the scientific literature. However, due to the scarcity of related datasets, previous NER attempts in this domain either suffer from low performance when deep learning based models are deployed, or they apply feature based machine learning models or rule based models to tackle this problem, which requires intensive efforts from domain experts, and limit the model generalization capability. In this paper, we propose EPICURE, an ensemble pre trained model equipped with a conditional random field pattern layer and a span prediction pattern layer to extract cancer mutations from text. We also adopt a data augmentation strategy to expand our training set from multiple datasets. Experimental results on three benchmark datasets show competitive results compared to the baseline models.
A systematic review of causal methods enabling predictions under hypothetical interventions
Nov 19, 2020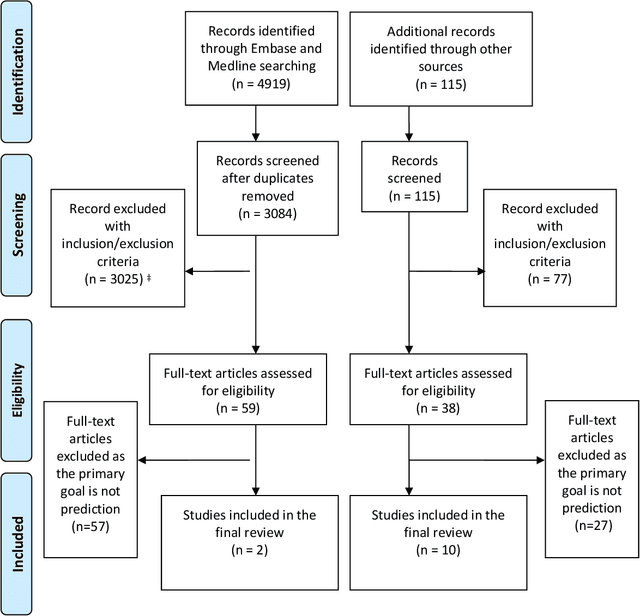

Abstract:Background: The methods with which prediction models are usually developed mean that neither the parameters nor the predictions should be interpreted causally. For many applications this is perfectly acceptable. However, when prediction models are used to support decision making, there is often a need for predicting outcomes under hypothetical interventions. Aims: We aimed to identify and compare published methods for developing and validating prediction models that enable risk estimation of outcomes under hypothetical interventions, utilizing causal inference. We aimed to identify the main methodological approaches, their underlying assumptions, targeted estimands, and possible sources of bias. Finally, we aimed to highlight unresolved methodological challenges. Methods: We systematically reviewed literature published by December 2019, considering papers in the health domain that used causal considerations to enable prediction models to be used to evaluate predictions under hypothetical interventions. We included both methodology development studies and applied studies. Results: We identified 4919 papers through database searches and a further 115 papers through manual searches. Of these, 87 papers were retained for full text screening, of which 12 were selected for inclusion. We found papers from both the statistical and the machine learning literature. Most of the identified methods for causal inference from observational data were based on marginal structural models and g-estimation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge