Vincent Jeanselme
One Loss to Rule Them All: Marked Time-to-Event for Structured EHR Foundation Models
Jan 31, 2026Abstract:Clinical events captured in Electronic Health Records (EHR) are irregularly sampled and may consist of a mixture of discrete events and numerical measurements, such as laboratory values or treatment dosages. The sequential nature of EHR, analogous to natural language, has motivated the use of next-token prediction to train prior EHR Foundation Models (FMs) over events. However, this training fails to capture the full structure of EHR. We propose ORA, a marked time-to-event pretraining objective that jointly models event timing and associated measurements. Across multiple datasets, downstream tasks, and model architectures, this objective consistently yields more generalizable representations than next-token prediction and pretraining losses that ignore continuous measurements. Importantly, the proposed objective yields improvements beyond traditional classification evaluation, including better regression and time-to-event prediction. Beyond introducing a new family of FMs, our results suggest a broader takeaway: pretraining objectives that account for EHR structure are critical for expanding downstream capabilities and generalizability
ADHAM: Additive Deep Hazard Analysis Mixtures for Interpretable Survival Regression
Sep 08, 2025Abstract:Survival analysis is a fundamental tool for modeling time-to-event outcomes in healthcare. Recent advances have introduced flexible neural network approaches for improved predictive performance. However, most of these models do not provide interpretable insights into the association between exposures and the modeled outcomes, a critical requirement for decision-making in clinical practice. To address this limitation, we propose Additive Deep Hazard Analysis Mixtures (ADHAM), an interpretable additive survival model. ADHAM assumes a conditional latent structure that defines subgroups, each characterized by a combination of covariate-specific hazard functions. To select the number of subgroups, we introduce a post-training refinement that reduces the number of equivalent latent subgroups by merging similar groups. We perform comprehensive studies to demonstrate ADHAM's interpretability at the population, subgroup, and individual levels. Extensive experiments on real-world datasets show that ADHAM provides novel insights into the association between exposures and outcomes. Further, ADHAM remains on par with existing state-of-the-art survival baselines in terms of predictive performance, offering a scalable and interpretable approach to time-to-event prediction in healthcare.
Prediction of Survival Outcomes under Clinical Presence Shift: A Joint Neural Network Architecture
Aug 07, 2025Abstract:Electronic health records arise from the complex interaction between patients and the healthcare system. This observation process of interactions, referred to as clinical presence, often impacts observed outcomes. When using electronic health records to develop clinical prediction models, it is standard practice to overlook clinical presence, impacting performance and limiting the transportability of models when this interaction evolves. We propose a multi-task recurrent neural network that jointly models the inter-observation time and the missingness processes characterising this interaction in parallel to the survival outcome of interest. Our work formalises the concept of clinical presence shift when the prediction model is deployed in new settings (e.g. different hospitals, regions or countries), and we theoretically justify why the proposed joint modelling can improve transportability under changes in clinical presence. We demonstrate, in a real-world mortality prediction task in the MIMIC-III dataset, how the proposed strategy improves performance and transportability compared to state-of-the-art prediction models that do not incorporate the observation process. These results emphasise the importance of leveraging clinical presence to improve performance and create more transportable clinical prediction models.
Competing Risks: Impact on Risk Estimation and Algorithmic Fairness
Aug 07, 2025Abstract:Accurate time-to-event prediction is integral to decision-making, informing medical guidelines, hiring decisions, and resource allocation. Survival analysis, the quantitative framework used to model time-to-event data, accounts for patients who do not experience the event of interest during the study period, known as censored patients. However, many patients experience events that prevent the observation of the outcome of interest. These competing risks are often treated as censoring, a practice frequently overlooked due to a limited understanding of its consequences. Our work theoretically demonstrates why treating competing risks as censoring introduces substantial bias in survival estimates, leading to systematic overestimation of risk and, critically, amplifying disparities. First, we formalize the problem of misclassifying competing risks as censoring and quantify the resulting error in survival estimates. Specifically, we develop a framework to estimate this error and demonstrate the associated implications for predictive performance and algorithmic fairness. Furthermore, we examine how differing risk profiles across demographic groups lead to group-specific errors, potentially exacerbating existing disparities. Our findings, supported by an empirical analysis of cardiovascular management, demonstrate that ignoring competing risks disproportionately impacts the individuals most at risk of these events, potentially accentuating inequity. By quantifying the error and highlighting the fairness implications of the common practice of considering competing risks as censoring, our work provides a critical insight into the development of survival models: practitioners must account for competing risks to improve accuracy, reduce disparities in risk assessment, and better inform downstream decisions.
ICYM2I: The illusion of multimodal informativeness under missingness
May 22, 2025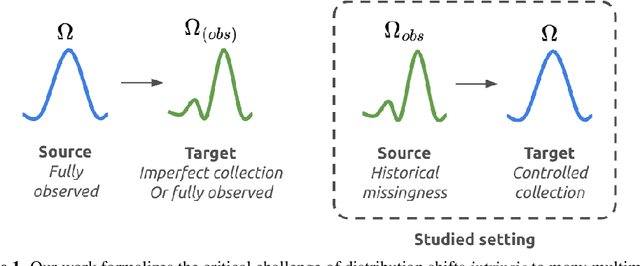
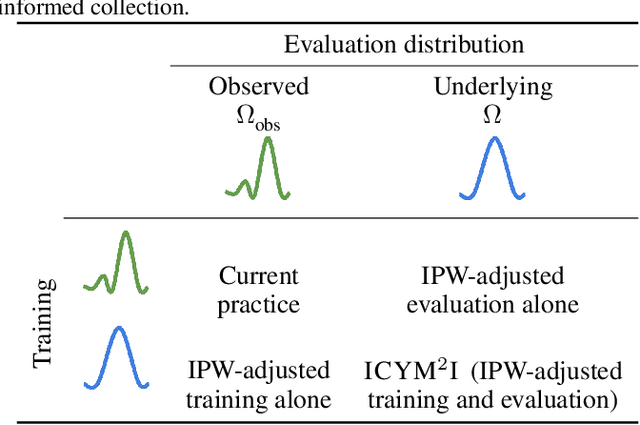
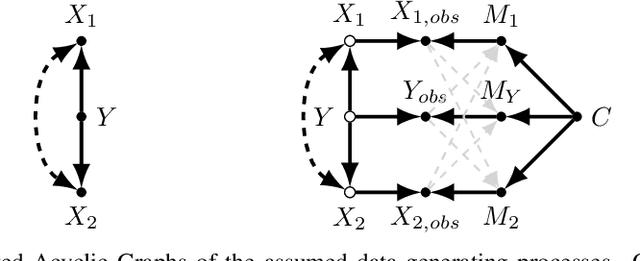
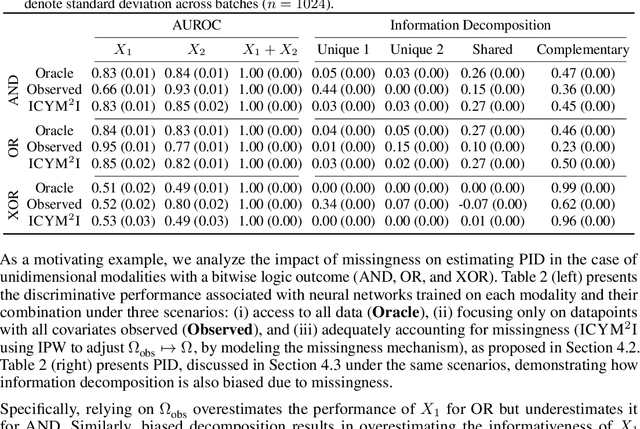
Abstract:Multimodal learning is of continued interest in artificial intelligence-based applications, motivated by the potential information gain from combining different types of data. However, modalities collected and curated during development may differ from the modalities available at deployment due to multiple factors including cost, hardware failure, or -- as we argue in this work -- the perceived informativeness of a given modality. Na{\"i}ve estimation of the information gain associated with including an additional modality without accounting for missingness may result in improper estimates of that modality's value in downstream tasks. Our work formalizes the problem of missingness in multimodal learning and demonstrates the biases resulting from ignoring this process. To address this issue, we introduce ICYM2I (In Case You Multimodal Missed It), a framework for the evaluation of predictive performance and information gain under missingness through inverse probability weighting-based correction. We demonstrate the importance of the proposed adjustment to estimate information gain under missingness on synthetic, semi-synthetic, and real-world medical datasets.
FoMoH: A clinically meaningful foundation model evaluation for structured electronic health records
May 22, 2025Abstract:Foundation models hold significant promise in healthcare, given their capacity to extract meaningful representations independent of downstream tasks. This property has enabled state-of-the-art performance across several clinical applications trained on structured electronic health record (EHR) data, even in settings with limited labeled data, a prevalent challenge in healthcare. However, there is little consensus on these models' potential for clinical utility due to the lack of desiderata of comprehensive and meaningful tasks and sufficiently diverse evaluations to characterize the benefit over conventional supervised learning. To address this gap, we propose a suite of clinically meaningful tasks spanning patient outcomes, early prediction of acute and chronic conditions, including desiderata for robust evaluations. We evaluate state-of-the-art foundation models on EHR data consisting of 5 million patients from Columbia University Irving Medical Center (CUMC), a large urban academic medical center in New York City, across 14 clinically relevant tasks. We measure overall accuracy, calibration, and subpopulation performance to surface tradeoffs based on the choice of pre-training, tokenization, and data representation strategies. Our study aims to advance the empirical evaluation of structured EHR foundation models and guide the development of future healthcare foundation models.
Identifying treatment response subgroups in observational time-to-event data
Aug 06, 2024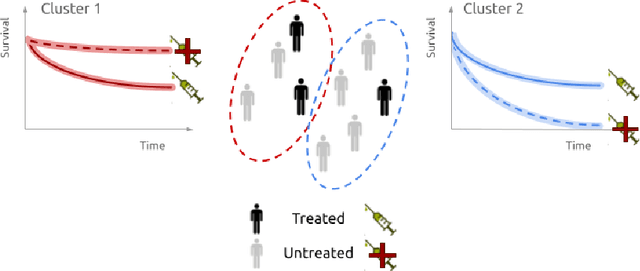
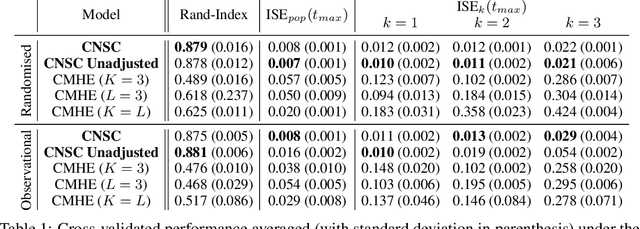
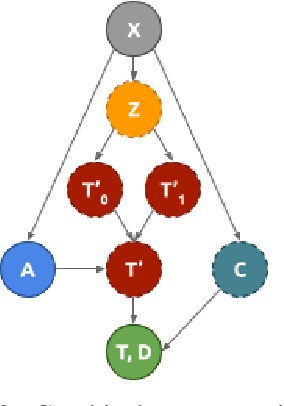
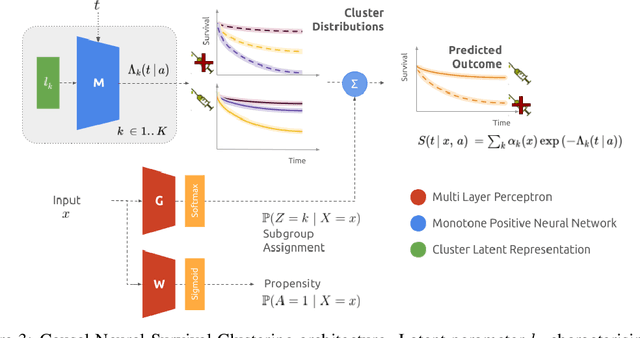
Abstract:Identifying patient subgroups with different treatment responses is an important task to inform medical recommendations, guidelines, and the design of future clinical trials. Existing approaches for subgroup analysis primarily focus on Randomised Controlled Trials (RCTs), in which treatment assignment is randomised. Furthermore, the patient cohort of an RCT is often constrained by cost, and is not representative of the heterogeneity of patients likely to receive treatment in real-world clinical practice. Therefore, when applied to observational studies, such approaches suffer from significant statistical biases because of the non-randomisation of treatment. Our work introduces a novel, outcome-guided method for identifying treatment response subgroups in observational studies. Our approach assigns each patient to a subgroup associated with two time-to-event distributions: one under treatment and one under control regime. It hence positions itself in between individualised and average treatment effect estimation. The assumptions of our model result in a simple correction of the statistical bias from treatment non-randomisation through inverse propensity weighting. In experiments, our approach significantly outperforms the current state-of-the-art method for outcome-guided subgroup analysis in both randomised and observational treatment regimes.
Recent Advances, Applications, and Open Challenges in Machine Learning for Health: Reflections from Research Roundtables at ML4H 2023 Symposium
Mar 03, 2024Abstract:The third ML4H symposium was held in person on December 10, 2023, in New Orleans, Louisiana, USA. The symposium included research roundtable sessions to foster discussions between participants and senior researchers on timely and relevant topics for the \ac{ML4H} community. Encouraged by the successful virtual roundtables in the previous year, we organized eleven in-person roundtables and four virtual roundtables at ML4H 2022. The organization of the research roundtables at the conference involved 17 Senior Chairs and 19 Junior Chairs across 11 tables. Each roundtable session included invited senior chairs (with substantial experience in the field), junior chairs (responsible for facilitating the discussion), and attendees from diverse backgrounds with interest in the session's topic. Herein we detail the organization process and compile takeaways from these roundtable discussions, including recent advances, applications, and open challenges for each topic. We conclude with a summary and lessons learned across all roundtables. This document serves as a comprehensive review paper, summarizing the recent advancements in machine learning for healthcare as contributed by foremost researchers in the field.
Neural Fine-Gray: Monotonic neural networks for competing risks
May 11, 2023Abstract:Time-to-event modelling, known as survival analysis, differs from standard regression as it addresses censoring in patients who do not experience the event of interest. Despite competitive performances in tackling this problem, machine learning methods often ignore other competing risks that preclude the event of interest. This practice biases the survival estimation. Extensions to address this challenge often rely on parametric assumptions or numerical estimations leading to sub-optimal survival approximations. This paper leverages constrained monotonic neural networks to model each competing survival distribution. This modelling choice ensures the exact likelihood maximisation at a reduced computational cost by using automatic differentiation. The effectiveness of the solution is demonstrated on one synthetic and three medical datasets. Finally, we discuss the implications of considering competing risks when developing risk scores for medical practice.
Imputation Strategies Under Clinical Presence: Impact on Algorithmic Fairness
Aug 13, 2022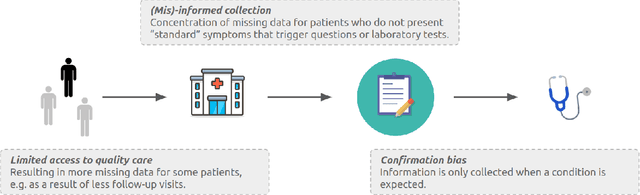
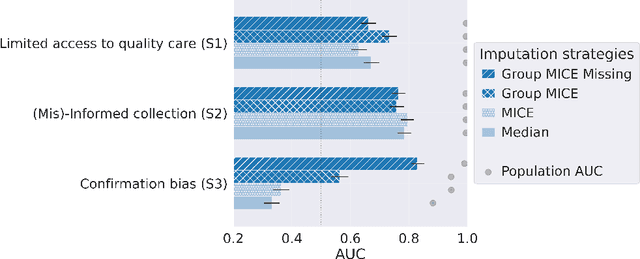
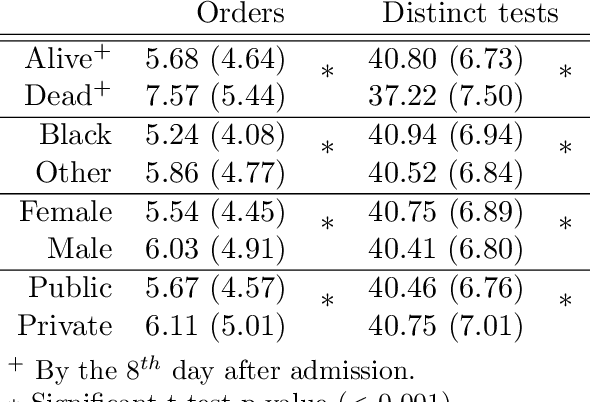
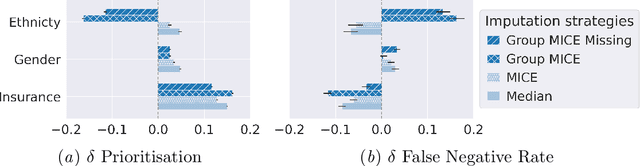
Abstract:Biases have marked medical history, leading to unequal care affecting marginalised groups. The patterns of missingness in observational data often reflect these group discrepancies, but the algorithmic fairness implications of group-specific missingness are not well understood. Despite its potential impact, imputation is too often a forgotten preprocessing step. At best, practitioners guide imputation choice by optimising overall performance, ignoring how this preprocessing can reinforce inequities. Our work questions this choice by studying how imputation affects downstream algorithmic fairness. First, we provide a structured view of the relationship between clinical presence mechanisms and group-specific missingness patterns. Then, through simulations and real-world experiments, we demonstrate that the imputation choice influences marginalised group performance and that no imputation strategy consistently reduces disparities. Importantly, our results show that current practices may endanger health equity as similarly performing imputation strategies at the population level can affect marginalised groups in different ways. Finally, we propose recommendations for mitigating inequity stemming from a neglected step of the machine learning pipeline.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge