Chao Pang
Columbia University Irving Medical Center
ERNIE 5.0 Technical Report
Feb 04, 2026Abstract:In this report, we introduce ERNIE 5.0, a natively autoregressive foundation model desinged for unified multimodal understanding and generation across text, image, video, and audio. All modalities are trained from scratch under a unified next-group-of-tokens prediction objective, based on an ultra-sparse mixture-of-experts (MoE) architecture with modality-agnostic expert routing. To address practical challenges in large-scale deployment under diverse resource constraints, ERNIE 5.0 adopts a novel elastic training paradigm. Within a single pre-training run, the model learns a family of sub-models with varying depths, expert capacities, and routing sparsity, enabling flexible trade-offs among performance, model size, and inference latency in memory- or time-constrained scenarios. Moreover, we systematically address the challenges of scaling reinforcement learning to unified foundation models, thereby guaranteeing efficient and stable post-training under ultra-sparse MoE architectures and diverse multimodal settings. Extensive experiments demonstrate that ERNIE 5.0 achieves strong and balanced performance across multiple modalities. To the best of our knowledge, among publicly disclosed models, ERNIE 5.0 represents the first production-scale realization of a trillion-parameter unified autoregressive model that supports both multimodal understanding and generation. To facilitate further research, we present detailed visualizations of modality-agnostic expert routing in the unified model, alongside comprehensive empirical analysis of elastic training, aiming to offer profound insights to the community.
One Loss to Rule Them All: Marked Time-to-Event for Structured EHR Foundation Models
Jan 31, 2026Abstract:Clinical events captured in Electronic Health Records (EHR) are irregularly sampled and may consist of a mixture of discrete events and numerical measurements, such as laboratory values or treatment dosages. The sequential nature of EHR, analogous to natural language, has motivated the use of next-token prediction to train prior EHR Foundation Models (FMs) over events. However, this training fails to capture the full structure of EHR. We propose ORA, a marked time-to-event pretraining objective that jointly models event timing and associated measurements. Across multiple datasets, downstream tasks, and model architectures, this objective consistently yields more generalizable representations than next-token prediction and pretraining losses that ignore continuous measurements. Importantly, the proposed objective yields improvements beyond traditional classification evaluation, including better regression and time-to-event prediction. Beyond introducing a new family of FMs, our results suggest a broader takeaway: pretraining objectives that account for EHR structure are critical for expanding downstream capabilities and generalizability
Toward Scalable Early Cancer Detection: Evaluating EHR-Based Predictive Models Against Traditional Screening Criteria
Nov 14, 2025Abstract:Current cancer screening guidelines cover only a few cancer types and rely on narrowly defined criteria such as age or a single risk factor like smoking history, to identify high-risk individuals. Predictive models using electronic health records (EHRs), which capture large-scale longitudinal patient-level health information, may provide a more effective tool for identifying high-risk groups by detecting subtle prediagnostic signals of cancer. Recent advances in large language and foundation models have further expanded this potential, yet evidence remains limited on how useful HER-based models are compared with traditional risk factors currently used in screening guidelines. We systematically evaluated the clinical utility of EHR-based predictive models against traditional risk factors, including gene mutations and family history of cancer, for identifying high-risk individuals across eight major cancers (breast, lung, colorectal, prostate, ovarian, liver, pancreatic, and stomach), using data from the All of Us Research Program, which integrates EHR, genomic, and survey data from over 865,000 participants. Even with a baseline modeling approach, EHR-based models achieved a 3- to 6-fold higher enrichment of true cancer cases among individuals identified as high risk compared with traditional risk factors alone, whether used as a standalone or complementary tool. The EHR foundation model, a state-of-the-art approach trained on comprehensive patient trajectories, further improved predictive performance across 26 cancer types, demonstrating the clinical potential of EHR-based predictive modeling to support more precise and scalable early detection strategies.
CEHR-GPT: A Scalable Multi-Task Foundation Model for Electronic Health Records
Sep 03, 2025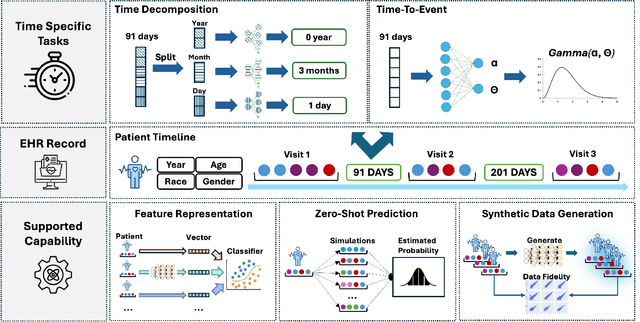


Abstract:Electronic Health Records (EHRs) provide a rich, longitudinal view of patient health and hold significant potential for advancing clinical decision support, risk prediction, and data-driven healthcare research. However, most artificial intelligence (AI) models for EHRs are designed for narrow, single-purpose tasks, limiting their generalizability and utility in real-world settings. Here, we present CEHR-GPT, a general-purpose foundation model for EHR data that unifies three essential capabilities - feature representation, zero-shot prediction, and synthetic data generation - within a single architecture. To support temporal reasoning over clinical sequences, \cehrgpt{} incorporates a novel time-token-based learning framework that explicitly encodes patients' dynamic timelines into the model structure. CEHR-GPT demonstrates strong performance across all three tasks and generalizes effectively to external datasets through vocabulary expansion and fine-tuning. Its versatility enables rapid model development, cohort discovery, and patient outcome forecasting without the need for task-specific retraining.
FoMoH: A clinically meaningful foundation model evaluation for structured electronic health records
May 22, 2025Abstract:Foundation models hold significant promise in healthcare, given their capacity to extract meaningful representations independent of downstream tasks. This property has enabled state-of-the-art performance across several clinical applications trained on structured electronic health record (EHR) data, even in settings with limited labeled data, a prevalent challenge in healthcare. However, there is little consensus on these models' potential for clinical utility due to the lack of desiderata of comprehensive and meaningful tasks and sufficiently diverse evaluations to characterize the benefit over conventional supervised learning. To address this gap, we propose a suite of clinically meaningful tasks spanning patient outcomes, early prediction of acute and chronic conditions, including desiderata for robust evaluations. We evaluate state-of-the-art foundation models on EHR data consisting of 5 million patients from Columbia University Irving Medical Center (CUMC), a large urban academic medical center in New York City, across 14 clinically relevant tasks. We measure overall accuracy, calibration, and subpopulation performance to surface tradeoffs based on the choice of pre-training, tokenization, and data representation strategies. Our study aims to advance the empirical evaluation of structured EHR foundation models and guide the development of future healthcare foundation models.
Vision-Language Modeling Meets Remote Sensing: Models, Datasets and Perspectives
May 20, 2025Abstract:Vision-language modeling (VLM) aims to bridge the information gap between images and natural language. Under the new paradigm of first pre-training on massive image-text pairs and then fine-tuning on task-specific data, VLM in the remote sensing domain has made significant progress. The resulting models benefit from the absorption of extensive general knowledge and demonstrate strong performance across a variety of remote sensing data analysis tasks. Moreover, they are capable of interacting with users in a conversational manner. In this paper, we aim to provide the remote sensing community with a timely and comprehensive review of the developments in VLM using the two-stage paradigm. Specifically, we first cover a taxonomy of VLM in remote sensing: contrastive learning, visual instruction tuning, and text-conditioned image generation. For each category, we detail the commonly used network architecture and pre-training objectives. Second, we conduct a thorough review of existing works, examining foundation models and task-specific adaptation methods in contrastive-based VLM, architectural upgrades, training strategies and model capabilities in instruction-based VLM, as well as generative foundation models with their representative downstream applications. Third, we summarize datasets used for VLM pre-training, fine-tuning, and evaluation, with an analysis of their construction methodologies (including image sources and caption generation) and key properties, such as scale and task adaptability. Finally, we conclude this survey with insights and discussions on future research directions: cross-modal representation alignment, vague requirement comprehension, explanation-driven model reliability, continually scalable model capabilities, and large-scale datasets featuring richer modalities and greater challenges.
SegEarth-R1: Geospatial Pixel Reasoning via Large Language Model
Apr 13, 2025Abstract:Remote sensing has become critical for understanding environmental dynamics, urban planning, and disaster management. However, traditional remote sensing workflows often rely on explicit segmentation or detection methods, which struggle to handle complex, implicit queries that require reasoning over spatial context, domain knowledge, and implicit user intent. Motivated by this, we introduce a new task, \ie, geospatial pixel reasoning, which allows implicit querying and reasoning and generates the mask of the target region. To advance this task, we construct and release the first large-scale benchmark dataset called EarthReason, which comprises 5,434 manually annotated image masks with over 30,000 implicit question-answer pairs. Moreover, we propose SegEarth-R1, a simple yet effective language-guided segmentation baseline that integrates a hierarchical visual encoder, a large language model (LLM) for instruction parsing, and a tailored mask generator for spatial correlation. The design of SegEarth-R1 incorporates domain-specific adaptations, including aggressive visual token compression to handle ultra-high-resolution remote sensing images, a description projection module to fuse language and multi-scale features, and a streamlined mask prediction pipeline that directly queries description embeddings. Extensive experiments demonstrate that SegEarth-R1 achieves state-of-the-art performance on both reasoning and referring segmentation tasks, significantly outperforming traditional and LLM-based segmentation methods. Our data and code will be released at https://github.com/earth-insights/SegEarth-R1.
Comparison of deep learning and conventional methods for disease onset prediction
Oct 14, 2024Abstract:Background: Conventional prediction methods such as logistic regression and gradient boosting have been widely utilized for disease onset prediction for their reliability and interpretability. Deep learning methods promise enhanced prediction performance by extracting complex patterns from clinical data, but face challenges like data sparsity and high dimensionality. Methods: This study compares conventional and deep learning approaches to predict lung cancer, dementia, and bipolar disorder using observational data from eleven databases from North America, Europe, and Asia. Models were developed using logistic regression, gradient boosting, ResNet, and Transformer, and validated both internally and externally across the data sources. Discrimination performance was assessed using AUROC, and calibration was evaluated using Eavg. Findings: Across 11 datasets, conventional methods generally outperformed deep learning methods in terms of discrimination performance, particularly during external validation, highlighting their better transportability. Learning curves suggest that deep learning models require substantially larger datasets to reach the same performance levels as conventional methods. Calibration performance was also better for conventional methods, with ResNet showing the poorest calibration. Interpretation: Despite the potential of deep learning models to capture complex patterns in structured observational healthcare data, conventional models remain highly competitive for disease onset prediction, especially in scenarios involving smaller datasets and if lengthy training times need to be avoided. The study underscores the need for future research focused on optimizing deep learning models to handle the sparsity, high dimensionality, and heterogeneity inherent in healthcare datasets, and find new strategies to exploit the full capabilities of deep learning methods.
H2RSVLM: Towards Helpful and Honest Remote Sensing Large Vision Language Model
Mar 29, 2024



Abstract:The generic large Vision-Language Models (VLMs) is rapidly developing, but still perform poorly in Remote Sensing (RS) domain, which is due to the unique and specialized nature of RS imagery and the comparatively limited spatial perception of current VLMs. Existing Remote Sensing specific Vision Language Models (RSVLMs) still have considerable potential for improvement, primarily owing to the lack of large-scale, high-quality RS vision-language datasets. We constructed HqDC-1.4M, the large scale High quality and Detailed Captions for RS images, containing 1.4 million image-caption pairs, which not only enhance the RSVLM's understanding of RS images but also significantly improve the model's spatial perception abilities, such as localization and counting, thereby increasing the helpfulness of the RSVLM. Moreover, to address the inevitable "hallucination" problem in RSVLM, we developed RSSA, the first dataset aimed at enhancing the Self-Awareness capability of RSVLMs. By incorporating a variety of unanswerable questions into typical RS visual question-answering tasks, RSSA effectively improves the truthfulness and reduces the hallucinations of the model's outputs, thereby enhancing the honesty of the RSVLM. Based on these datasets, we proposed the H2RSVLM, the Helpful and Honest Remote Sensing Vision Language Model. H2RSVLM has achieved outstanding performance on multiple RS public datasets and is capable of recognizing and refusing to answer the unanswerable questions, effectively mitigating the incorrect generations. We will release the code, data and model weights at https://github.com/opendatalab/H2RSVLM .
CEHR-GPT: Generating Electronic Health Records with Chronological Patient Timelines
Feb 06, 2024



Abstract:Synthetic Electronic Health Records (EHR) have emerged as a pivotal tool in advancing healthcare applications and machine learning models, particularly for researchers without direct access to healthcare data. Although existing methods, like rule-based approaches and generative adversarial networks (GANs), generate synthetic data that resembles real-world EHR data, these methods often use a tabular format, disregarding temporal dependencies in patient histories and limiting data replication. Recently, there has been a growing interest in leveraging Generative Pre-trained Transformers (GPT) for EHR data. This enables applications like disease progression analysis, population estimation, counterfactual reasoning, and synthetic data generation. In this work, we focus on synthetic data generation and demonstrate the capability of training a GPT model using a particular patient representation derived from CEHR-BERT, enabling us to generate patient sequences that can be seamlessly converted to the Observational Medical Outcomes Partnership (OMOP) data format.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge