Jenna M. Reps
Clinical semantics for lung cancer prediction
Aug 20, 2025Abstract:Background: Existing clinical prediction models often represent patient data using features that ignore the semantic relationships between clinical concepts. This study integrates domain-specific semantic information by mapping the SNOMED medical term hierarchy into a low-dimensional hyperbolic space using Poincar\'e embeddings, with the aim of improving lung cancer onset prediction. Methods: Using a retrospective cohort from the Optum EHR dataset, we derived a clinical knowledge graph from the SNOMED taxonomy and generated Poincar\'e embeddings via Riemannian stochastic gradient descent. These embeddings were then incorporated into two deep learning architectures, a ResNet and a Transformer model. Models were evaluated for discrimination (area under the receiver operating characteristic curve) and calibration (average absolute difference between observed and predicted probabilities) performance. Results: Incorporating pre-trained Poincar\'e embeddings resulted in modest and consistent improvements in discrimination performance compared to baseline models using randomly initialized Euclidean embeddings. ResNet models, particularly those using a 10-dimensional Poincar\'e embedding, showed enhanced calibration, whereas Transformer models maintained stable calibration across configurations. Discussion: Embedding clinical knowledge graphs into hyperbolic space and integrating these representations into deep learning models can improve lung cancer onset prediction by preserving the hierarchical structure of clinical terminologies used for prediction. This approach demonstrates a feasible method for combining data-driven feature extraction with established clinical knowledge.
Comparison of deep learning and conventional methods for disease onset prediction
Oct 14, 2024Abstract:Background: Conventional prediction methods such as logistic regression and gradient boosting have been widely utilized for disease onset prediction for their reliability and interpretability. Deep learning methods promise enhanced prediction performance by extracting complex patterns from clinical data, but face challenges like data sparsity and high dimensionality. Methods: This study compares conventional and deep learning approaches to predict lung cancer, dementia, and bipolar disorder using observational data from eleven databases from North America, Europe, and Asia. Models were developed using logistic regression, gradient boosting, ResNet, and Transformer, and validated both internally and externally across the data sources. Discrimination performance was assessed using AUROC, and calibration was evaluated using Eavg. Findings: Across 11 datasets, conventional methods generally outperformed deep learning methods in terms of discrimination performance, particularly during external validation, highlighting their better transportability. Learning curves suggest that deep learning models require substantially larger datasets to reach the same performance levels as conventional methods. Calibration performance was also better for conventional methods, with ResNet showing the poorest calibration. Interpretation: Despite the potential of deep learning models to capture complex patterns in structured observational healthcare data, conventional models remain highly competitive for disease onset prediction, especially in scenarios involving smaller datasets and if lengthy training times need to be avoided. The study underscores the need for future research focused on optimizing deep learning models to handle the sparsity, high dimensionality, and heterogeneity inherent in healthcare datasets, and find new strategies to exploit the full capabilities of deep learning methods.
How little data do we need for patient-level prediction?
Aug 14, 2020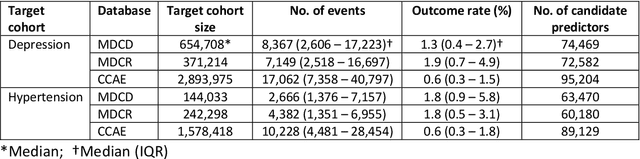


Abstract:Objective: Provide guidance on sample size considerations for developing predictive models by empirically establishing the adequate sample size, which balances the competing objectives of improving model performance and reducing model complexity as well as computational requirements. Materials and Methods: We empirically assess the effect of sample size on prediction performance and model complexity by generating learning curves for 81 prediction problems in three large observational health databases, requiring training of 17,248 prediction models. The adequate sample size was defined as the sample size for which the performance of a model equalled the maximum model performance minus a small threshold value. Results: The adequate sample size achieves a median reduction of the number of observations between 9.5% and 78.5% for threshold values between 0.001 and 0.02. The median reduction of the number of predictors in the models at the adequate sample size varied between 8.6% and 68.3%, respectively. Discussion: Based on our results a conservative, yet significant, reduction in sample size and model complexity can be estimated for future prediction work. Though, if a researcher is willing to generate a learning curve a much larger reduction of the model complexity may be possible as suggested by a large outcome-dependent variability. Conclusion: Our results suggest that in most cases only a fraction of the available data was sufficient to produce a model close to the performance of one developed on the full data set, but with a substantially reduced model complexity.
Refining adverse drug reaction signals by incorporating interaction variables identified using emergent pattern mining
Jul 20, 2016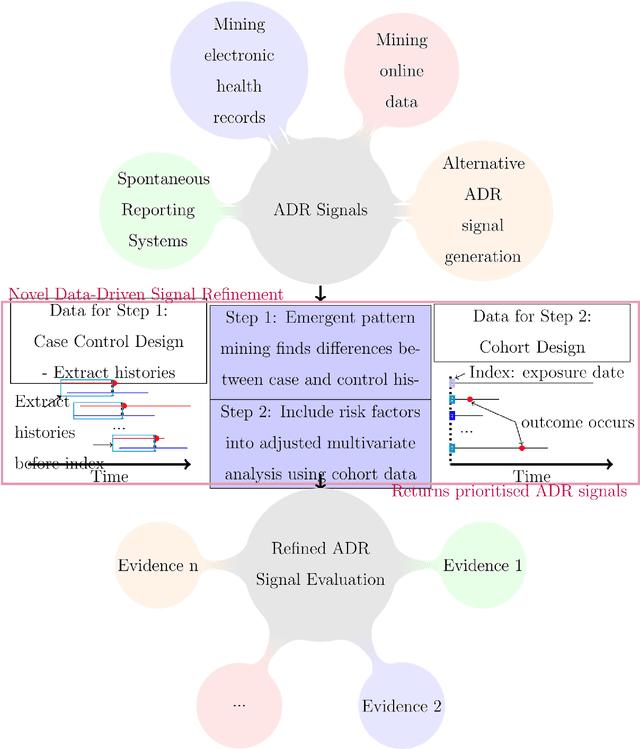
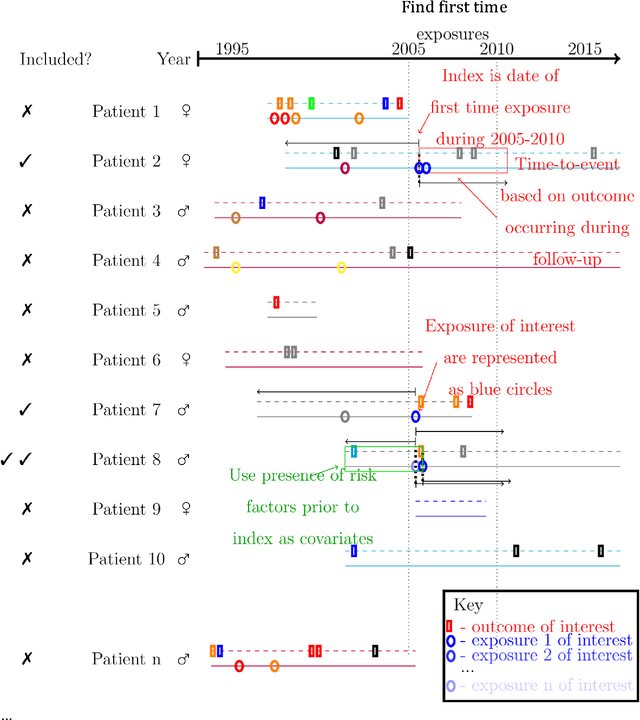
Abstract:Purpose: To develop a framework for identifying and incorporating candidate confounding interaction terms into a regularised cox regression analysis to refine adverse drug reaction signals obtained via longitudinal observational data. Methods: We considered six drug families that are commonly associated with myocardial infarction in observational healthcare data, but where the causal relationship ground truth is known (adverse drug reaction or not). We applied emergent pattern mining to find itemsets of drugs and medical events that are associated with the development of myocardial infarction. These are the candidate confounding interaction terms. We then implemented a cohort study design using regularised cox regression that incorporated and accounted for the candidate confounding interaction terms. Results The methodology was able to account for signals generated due to confounding and a cox regression with elastic net regularisation correctly ranked the drug families known to be true adverse drug reactions above those.
Refining Adverse Drug Reactions using Association Rule Mining for Electronic Healthcare Data
Feb 20, 2015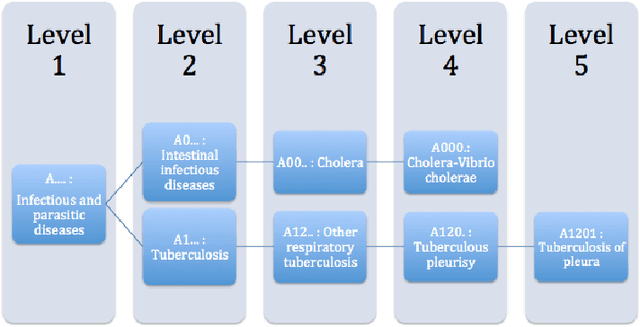
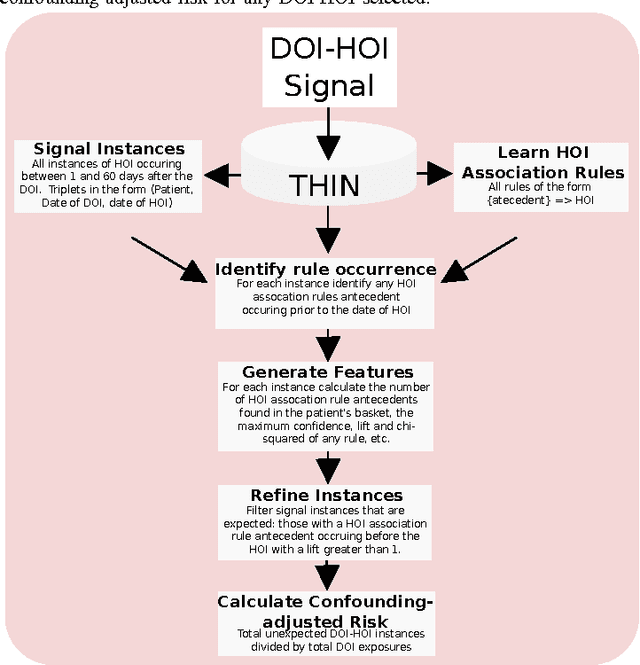
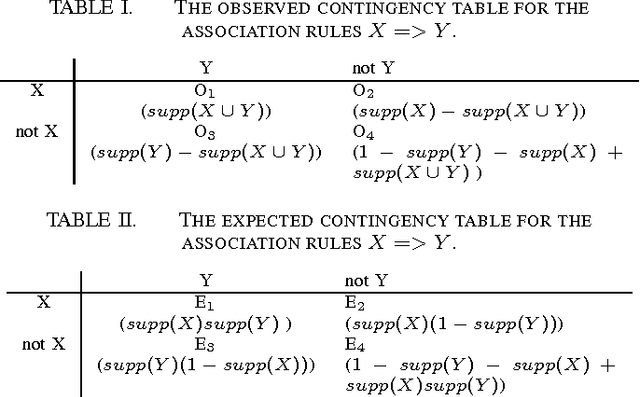

Abstract:Side effects of prescribed medications are a common occurrence. Electronic healthcare databases present the opportunity to identify new side effects efficiently but currently the methods are limited due to confounding (i.e. when an association between two variables is identified due to them both being associated to a third variable). In this paper we propose a proof of concept method that learns common associations and uses this knowledge to automatically refine side effect signals (i.e. exposure-outcome associations) by removing instances of the exposure-outcome associations that are caused by confounding. This leaves the signal instances that are most likely to correspond to true side effect occurrences. We then calculate a novel measure termed the confounding-adjusted risk value, a more accurate absolute risk value of a patient experiencing the outcome within 60 days of the exposure. Tentative results suggest that the method works. For the four signals (i.e. exposure-outcome associations) investigated we are able to correctly filter the majority of exposure-outcome instances that were unlikely to correspond to true side effects. The method is likely to improve when tuning the association rule mining parameters for specific health outcomes. This paper shows that it may be possible to filter signals at a patient level based on association rules learned from considering patients' medical histories. However, additional work is required to develop a way to automate the tuning of the method's parameters.
Tuning a Multiple Classifier System for Side Effect Discovery using Genetic Algorithms
Sep 03, 2014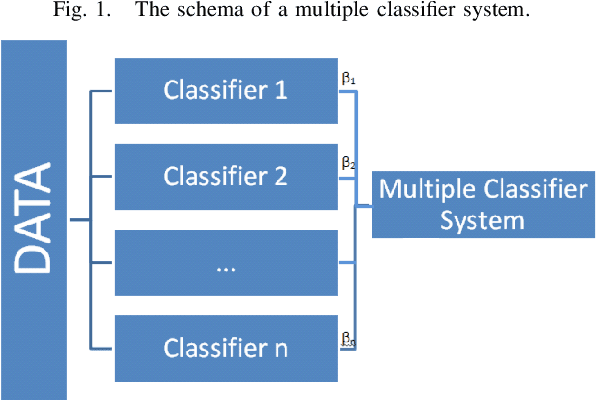
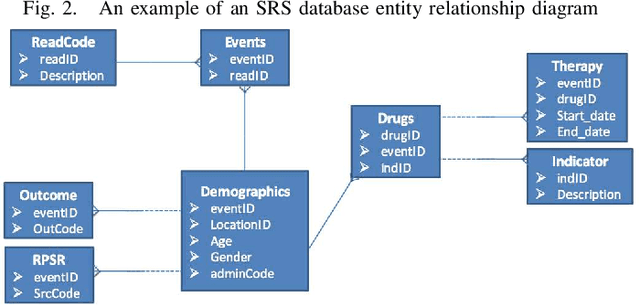
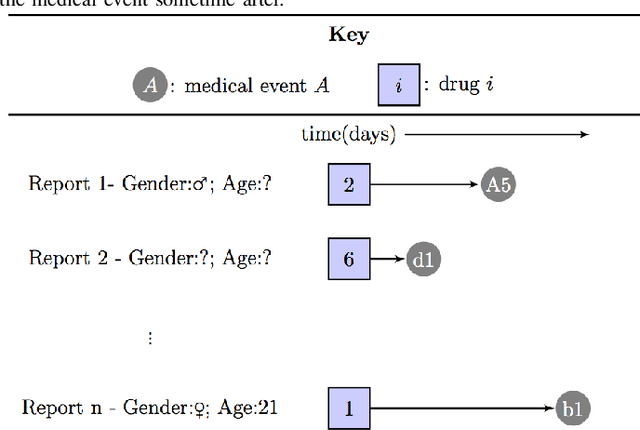

Abstract:In previous work, a novel supervised framework implementing a binary classifier was presented that obtained excellent results for side effect discovery. Interestingly, unique side effects were identified when different binary classifiers were used within the framework, prompting the investigation of applying a multiple classifier system. In this paper we investigate tuning a side effect multiple classifying system using genetic algorithms. The results of this research show that the novel framework implementing a multiple classifying system trained using genetic algorithms can obtain a higher partial area under the receiver operating characteristic curve than implementing a single classifier. Furthermore, the framework is able to detect side effects efficiently and obtains a low false positive rate.
Signalling Paediatric Side Effects using an Ensemble of Simple Study Designs
Sep 02, 2014
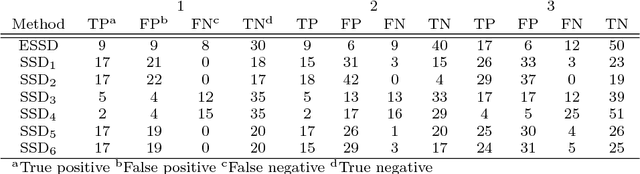
Abstract:Background: Children are frequently prescribed medication off-label, meaning there has not been sufficient testing of the medication to determine its safety or effectiveness. The main reason this safety knowledge is lacking is due to ethical restrictions that prevent children from being included in the majority of clinical trials. Objective: The objective of this paper is to investigate whether an ensemble of simple study designs can be implemented to signal acutely occurring side effects effectively within the paediatric population by using historical longitudinal data. The majority of pharmacovigilance techniques are unsupervised, but this research presents a supervised framework. Methods: Multiple measures of association are calculated for each drug and medical event pair and these are used as features that are fed into a classiffier to determine the likelihood of the drug and medical event pair corresponding to an adverse drug reaction. The classiffier is trained using known adverse drug reactions or known non-adverse drug reaction relationships. Results: The novel ensemble framework obtained a false positive rate of 0:149, a sensitivity of 0:547 and a specificity of 0:851 when implemented on a reference set of drug and medical event pairs. The novel framework consistently outperformed each individual simple study design. Conclusion: This research shows that it is possible to exploit the mechanism of causality and presents a framework for signalling adverse drug reactions effectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge