Patrick B. Ryan
An Open-Source Knowledge Graph Ecosystem for the Life Sciences
Jul 11, 2023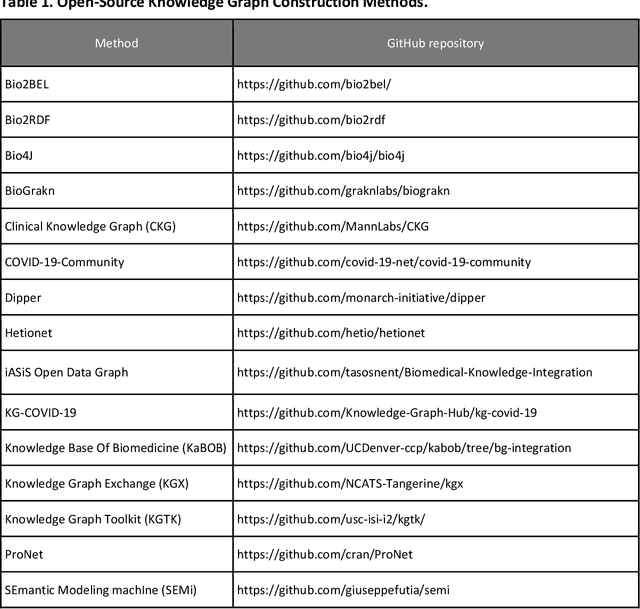
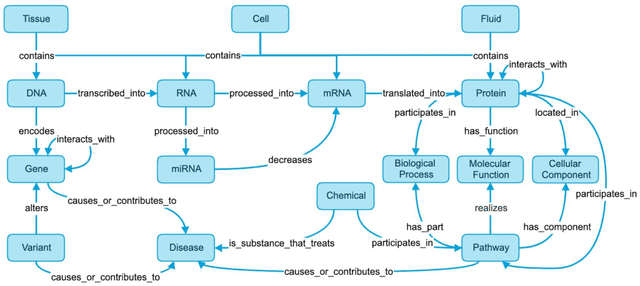
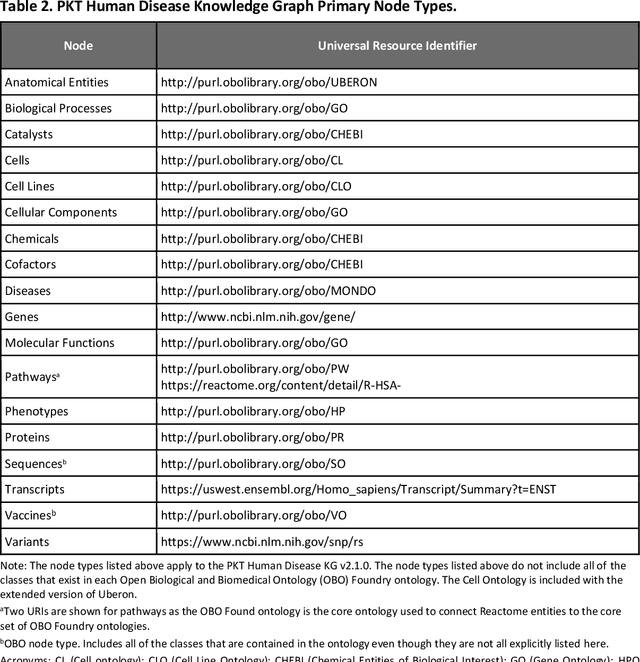
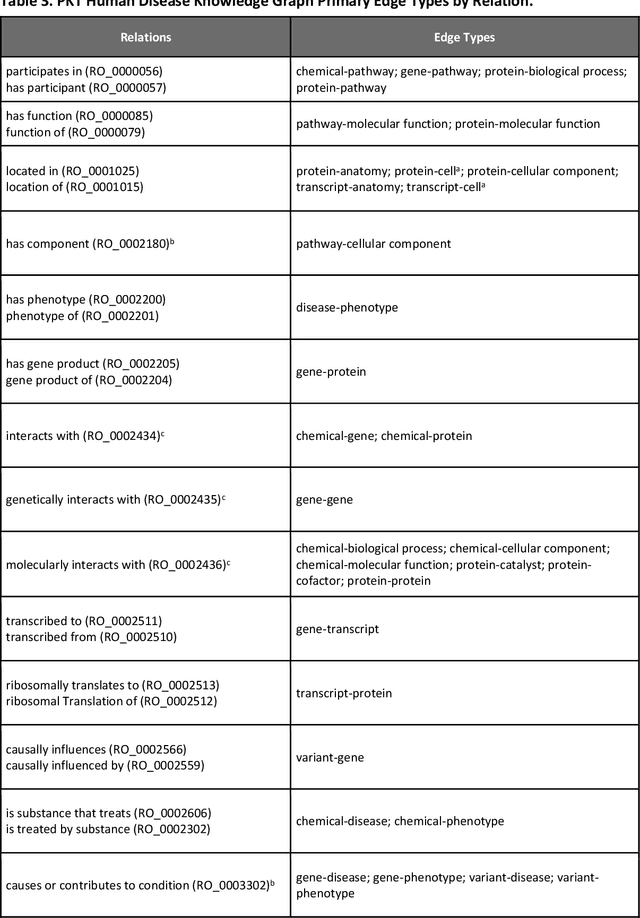
Abstract:Translational research requires data at multiple scales of biological organization. Advancements in sequencing and multi-omics technologies have increased the availability of these data but researchers face significant integration challenges. Knowledge graphs (KGs) are used to model complex phenomena, and methods exist to automatically construct them. However, tackling complex biomedical integration problems requires flexibility in the way knowledge is modeled. Moreover, existing KG construction methods provide robust tooling at the cost of fixed or limited choices among knowledge representation models. PheKnowLator (Phenotype Knowledge Translator) is a semantic ecosystem for automating the FAIR (Findable, Accessible, Interoperable, and Reusable) construction of ontologically grounded KGs with fully customizable knowledge representation. The ecosystem includes KG construction resources (e.g., data preparation APIs), analysis tools (e.g., SPARQL endpoints and abstraction algorithms), and benchmarks (e.g., prebuilt KGs and embeddings). We evaluate the ecosystem by surveying open-source KG construction methods and analyzing its computational performance when constructing 12 large-scale KGs. With flexible knowledge representation, PheKnowLator enables fully customizable KGs without compromising performance or usability.
A standardized framework for risk-based assessment of treatment effect heterogeneity in observational healthcare databases
Oct 13, 2020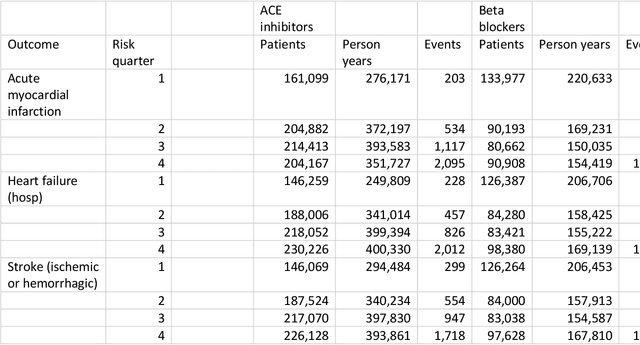
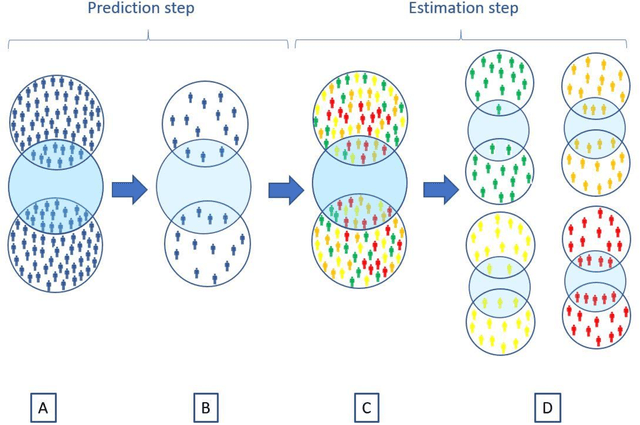
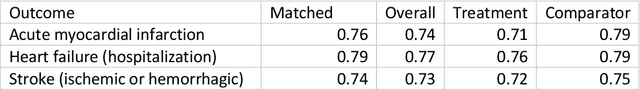
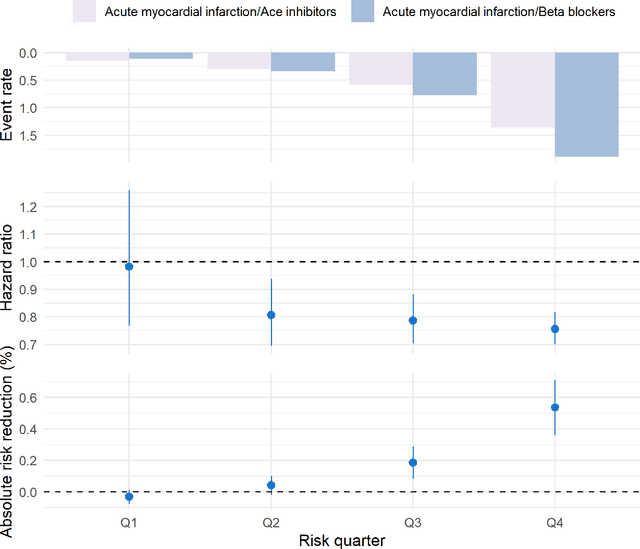
Abstract:Aim: One of the aims of the Observation Health Data Sciences and Informatics (OHDSI) initiative is population-level treatment effect estimation in large observational databases. Since treatment effects are well-known to vary across groups of patients with different baseline risk, we aimed to extend the OHDSI methods library with a framework for risk-based assessment of treatment effect heterogeneity. Materials and Methods: The proposed framework consists of five steps: 1) definition of the problem, i.e. the population, the treatment, the comparator and the outcome(s) of interest; 2) identification of relevant databases; 3) development of a prediction model for the outcome(s) of interest; 4) estimation of propensity scores within strata of predicted risk and estimation of relative and absolute treatment effect within strata of predicted risk; 5) evaluation and presentation of results. Results: We demonstrate our framework by evaluating heterogeneity of the effect of angiotensin-converting enzyme (ACE) inhibitors versus beta blockers on a set of 9 outcomes of interest across three observational databases. With increasing risk of acute myocardial infarction we observed increasing absolute benefits, i.e. from -0.03% to 0.54% in the lowest to highest risk groups. Cough-related absolute harms decreased from 4.1% to 2.6%. Conclusions: The proposed framework may be useful for the evaluation of heterogeneity of treatment effect on observational data that are mapped to the OMOP Common Data Model. The proof of concept study demonstrates its feasibility in large observational data. Further insights may arise by application to safety and effectiveness questions across the global data network.
How little data do we need for patient-level prediction?
Aug 14, 2020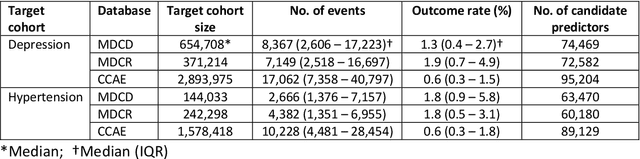


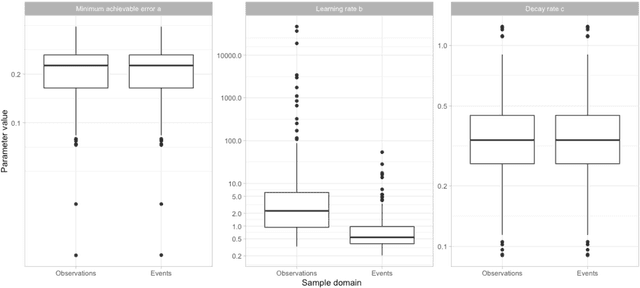
Abstract:Objective: Provide guidance on sample size considerations for developing predictive models by empirically establishing the adequate sample size, which balances the competing objectives of improving model performance and reducing model complexity as well as computational requirements. Materials and Methods: We empirically assess the effect of sample size on prediction performance and model complexity by generating learning curves for 81 prediction problems in three large observational health databases, requiring training of 17,248 prediction models. The adequate sample size was defined as the sample size for which the performance of a model equalled the maximum model performance minus a small threshold value. Results: The adequate sample size achieves a median reduction of the number of observations between 9.5% and 78.5% for threshold values between 0.001 and 0.02. The median reduction of the number of predictors in the models at the adequate sample size varied between 8.6% and 68.3%, respectively. Discussion: Based on our results a conservative, yet significant, reduction in sample size and model complexity can be estimated for future prediction work. Though, if a researcher is willing to generate a learning curve a much larger reduction of the model complexity may be possible as suggested by a large outcome-dependent variability. Conclusion: Our results suggest that in most cases only a fraction of the available data was sufficient to produce a model close to the performance of one developed on the full data set, but with a substantially reduced model complexity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge