Shalmali Joshi
One Loss to Rule Them All: Marked Time-to-Event for Structured EHR Foundation Models
Jan 31, 2026Abstract:Clinical events captured in Electronic Health Records (EHR) are irregularly sampled and may consist of a mixture of discrete events and numerical measurements, such as laboratory values or treatment dosages. The sequential nature of EHR, analogous to natural language, has motivated the use of next-token prediction to train prior EHR Foundation Models (FMs) over events. However, this training fails to capture the full structure of EHR. We propose ORA, a marked time-to-event pretraining objective that jointly models event timing and associated measurements. Across multiple datasets, downstream tasks, and model architectures, this objective consistently yields more generalizable representations than next-token prediction and pretraining losses that ignore continuous measurements. Importantly, the proposed objective yields improvements beyond traditional classification evaluation, including better regression and time-to-event prediction. Beyond introducing a new family of FMs, our results suggest a broader takeaway: pretraining objectives that account for EHR structure are critical for expanding downstream capabilities and generalizability
A pipeline for enabling path-specific causal fairness in observational health data
Jan 14, 2026Abstract:When training machine learning (ML) models for potential deployment in a healthcare setting, it is essential to ensure that they do not replicate or exacerbate existing healthcare biases. Although many definitions of fairness exist, we focus on path-specific causal fairness, which allows us to better consider the social and medical contexts in which biases occur (e.g., direct discrimination by a clinician or model versus bias due to differential access to the healthcare system) and to characterize how these biases may appear in learned models. In this work, we map the structural fairness model to the observational healthcare setting and create a generalizable pipeline for training causally fair models. The pipeline explicitly considers specific healthcare context and disparities to define a target "fair" model. Our work fills two major gaps: first, we expand on characterizations of the "fairness-accuracy" tradeoff by detangling direct and indirect sources of bias and jointly presenting these fairness considerations alongside considerations of accuracy in the context of broadly known biases. Second, we demonstrate how a foundation model trained without fairness constraints on observational health data can be leveraged to generate causally fair downstream predictions in tasks with known social and medical disparities. This work presents a model-agnostic pipeline for training causally fair machine learning models that address both direct and indirect forms of healthcare bias.
Learning-To-Measure: In-context Active Feature Acquisition
Oct 14, 2025



Abstract:Active feature acquisition (AFA) is a sequential decision-making problem where the goal is to improve model performance for test instances by adaptively selecting which features to acquire. In practice, AFA methods often learn from retrospective data with systematic missingness in the features and limited task-specific labels. Most prior work addresses acquisition for a single predetermined task, limiting scalability. To address this limitation, we formalize the meta-AFA problem, where the goal is to learn acquisition policies across various tasks. We introduce Learning-to-Measure (L2M), which consists of i) reliable uncertainty quantification over unseen tasks, and ii) an uncertainty-guided greedy feature acquisition agent that maximizes conditional mutual information. We demonstrate a sequence-modeling or autoregressive pre-training approach that underpins reliable uncertainty quantification for tasks with arbitrary missingness. L2M operates directly on datasets with retrospective missingness and performs the meta-AFA task in-context, eliminating per-task retraining. Across synthetic and real-world tabular benchmarks, L2M matches or surpasses task-specific baselines, particularly under scarce labels and high missingness.
ADHAM: Additive Deep Hazard Analysis Mixtures for Interpretable Survival Regression
Sep 08, 2025Abstract:Survival analysis is a fundamental tool for modeling time-to-event outcomes in healthcare. Recent advances have introduced flexible neural network approaches for improved predictive performance. However, most of these models do not provide interpretable insights into the association between exposures and the modeled outcomes, a critical requirement for decision-making in clinical practice. To address this limitation, we propose Additive Deep Hazard Analysis Mixtures (ADHAM), an interpretable additive survival model. ADHAM assumes a conditional latent structure that defines subgroups, each characterized by a combination of covariate-specific hazard functions. To select the number of subgroups, we introduce a post-training refinement that reduces the number of equivalent latent subgroups by merging similar groups. We perform comprehensive studies to demonstrate ADHAM's interpretability at the population, subgroup, and individual levels. Extensive experiments on real-world datasets show that ADHAM provides novel insights into the association between exposures and outcomes. Further, ADHAM remains on par with existing state-of-the-art survival baselines in terms of predictive performance, offering a scalable and interpretable approach to time-to-event prediction in healthcare.
CEHR-GPT: A Scalable Multi-Task Foundation Model for Electronic Health Records
Sep 03, 2025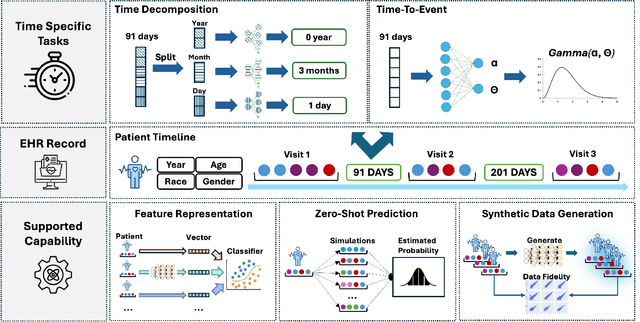


Abstract:Electronic Health Records (EHRs) provide a rich, longitudinal view of patient health and hold significant potential for advancing clinical decision support, risk prediction, and data-driven healthcare research. However, most artificial intelligence (AI) models for EHRs are designed for narrow, single-purpose tasks, limiting their generalizability and utility in real-world settings. Here, we present CEHR-GPT, a general-purpose foundation model for EHR data that unifies three essential capabilities - feature representation, zero-shot prediction, and synthetic data generation - within a single architecture. To support temporal reasoning over clinical sequences, \cehrgpt{} incorporates a novel time-token-based learning framework that explicitly encodes patients' dynamic timelines into the model structure. CEHR-GPT demonstrates strong performance across all three tasks and generalizes effectively to external datasets through vocabulary expansion and fine-tuning. Its versatility enables rapid model development, cohort discovery, and patient outcome forecasting without the need for task-specific retraining.
Path-specific effects for pulse-oximetry guided decisions in critical care
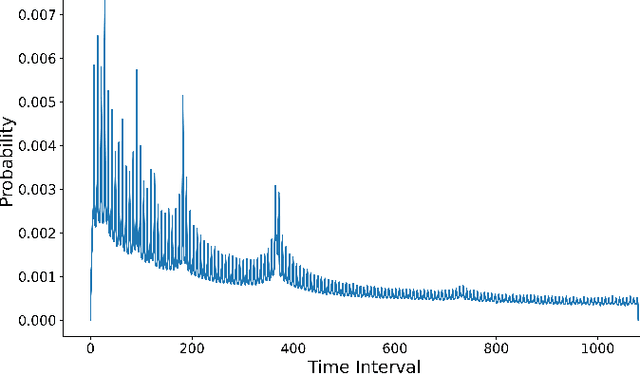
Jun 14, 2025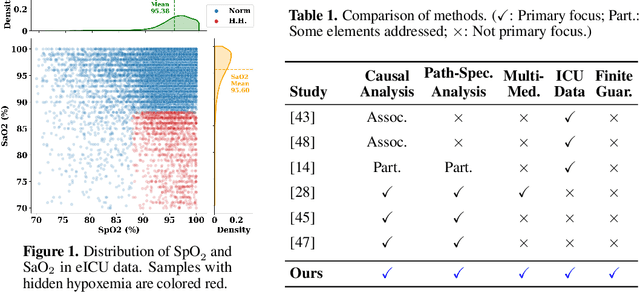
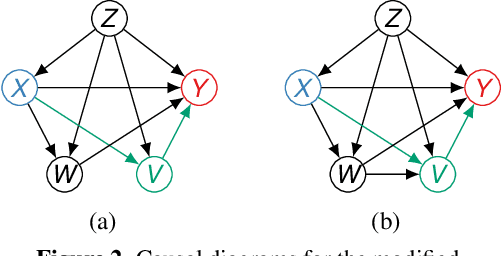
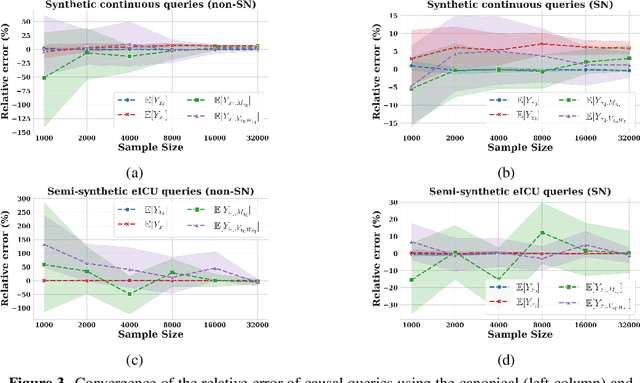
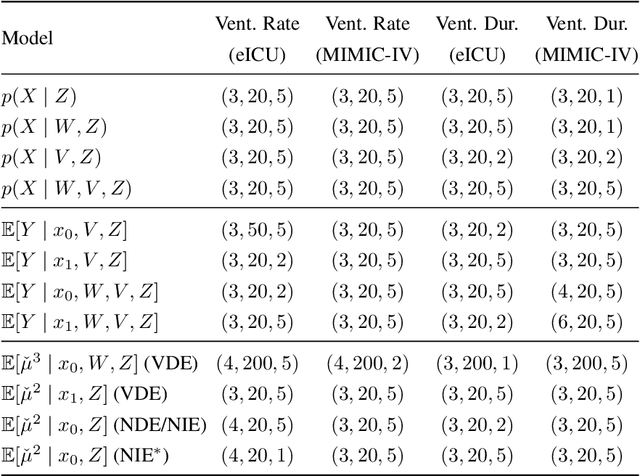
Abstract:Identifying and measuring biases associated with sensitive attributes is a crucial consideration in healthcare to prevent treatment disparities. One prominent issue is inaccurate pulse oximeter readings, which tend to overestimate oxygen saturation for dark-skinned patients and misrepresent supplemental oxygen needs. Most existing research has revealed statistical disparities linking device errors to patient outcomes in intensive care units (ICUs) without causal formalization. In contrast, this study causally investigates how racial discrepancies in oximetry measurements affect invasive ventilation in ICU settings. We employ a causal inference-based approach using path-specific effects to isolate the impact of bias by race on clinical decision-making. To estimate these effects, we leverage a doubly robust estimator, propose its self-normalized variant for improved sample efficiency, and provide novel finite-sample guarantees. Our methodology is validated on semi-synthetic data and applied to two large real-world health datasets: MIMIC-IV and eICU. Contrary to prior work, our analysis reveals minimal impact of racial discrepancies on invasive ventilation rates. However, path-specific effects mediated by oxygen saturation disparity are more pronounced on ventilation duration, and the severity differs by dataset. Our work provides a novel and practical pipeline for investigating potential disparities in the ICU and, more crucially, highlights the necessity of causal methods to robustly assess fairness in decision-making.
ICYM2I: The illusion of multimodal informativeness under missingness
May 22, 2025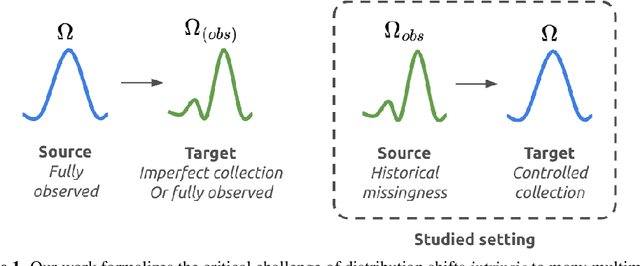
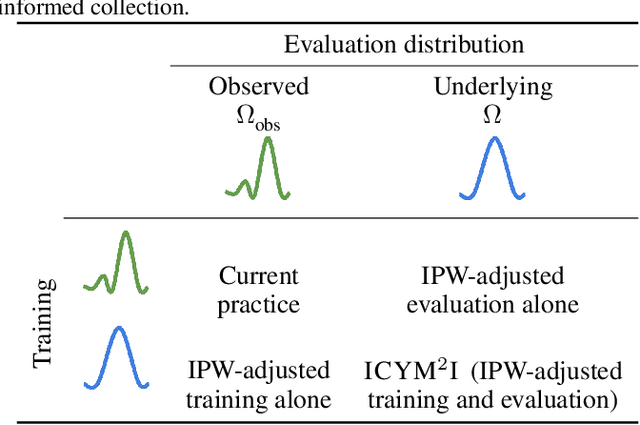
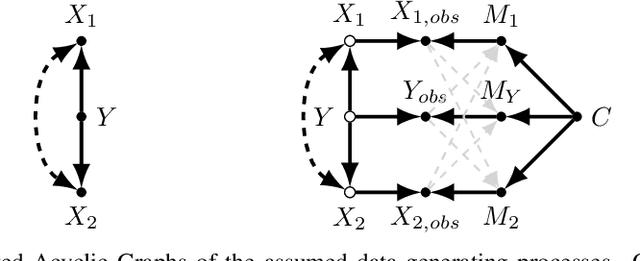
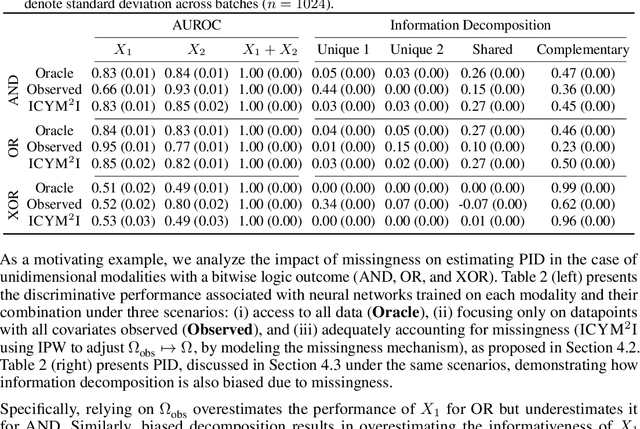
Abstract:Multimodal learning is of continued interest in artificial intelligence-based applications, motivated by the potential information gain from combining different types of data. However, modalities collected and curated during development may differ from the modalities available at deployment due to multiple factors including cost, hardware failure, or -- as we argue in this work -- the perceived informativeness of a given modality. Na{\"i}ve estimation of the information gain associated with including an additional modality without accounting for missingness may result in improper estimates of that modality's value in downstream tasks. Our work formalizes the problem of missingness in multimodal learning and demonstrates the biases resulting from ignoring this process. To address this issue, we introduce ICYM2I (In Case You Multimodal Missed It), a framework for the evaluation of predictive performance and information gain under missingness through inverse probability weighting-based correction. We demonstrate the importance of the proposed adjustment to estimate information gain under missingness on synthetic, semi-synthetic, and real-world medical datasets.
FoMoH: A clinically meaningful foundation model evaluation for structured electronic health records
May 22, 2025Abstract:Foundation models hold significant promise in healthcare, given their capacity to extract meaningful representations independent of downstream tasks. This property has enabled state-of-the-art performance across several clinical applications trained on structured electronic health record (EHR) data, even in settings with limited labeled data, a prevalent challenge in healthcare. However, there is little consensus on these models' potential for clinical utility due to the lack of desiderata of comprehensive and meaningful tasks and sufficiently diverse evaluations to characterize the benefit over conventional supervised learning. To address this gap, we propose a suite of clinically meaningful tasks spanning patient outcomes, early prediction of acute and chronic conditions, including desiderata for robust evaluations. We evaluate state-of-the-art foundation models on EHR data consisting of 5 million patients from Columbia University Irving Medical Center (CUMC), a large urban academic medical center in New York City, across 14 clinically relevant tasks. We measure overall accuracy, calibration, and subpopulation performance to surface tradeoffs based on the choice of pre-training, tokenization, and data representation strategies. Our study aims to advance the empirical evaluation of structured EHR foundation models and guide the development of future healthcare foundation models.
A Planning Framework for Adaptive Labeling
Feb 10, 2025Abstract:Ground truth labels/outcomes are critical for advancing scientific and engineering applications, e.g., evaluating the treatment effect of an intervention or performance of a predictive model. Since randomly sampling inputs for labeling can be prohibitively expensive, we introduce an adaptive labeling framework where measurement effort can be reallocated in batches. We formulate this problem as a Markov decision process where posterior beliefs evolve over time as batches of labels are collected (state transition), and batches (actions) are chosen to minimize uncertainty at the end of data collection. We design a computational framework that is agnostic to different uncertainty quantification approaches including those based on deep learning, and allows a diverse array of policy gradient approaches by relying on continuous policy parameterizations. On real and synthetic datasets, we demonstrate even a one-step lookahead policy can substantially outperform common adaptive labeling heuristics, highlighting the virtue of planning. On the methodological side, we note that standard REINFORCE-style policy gradient estimators can suffer high variance since they rely only on zeroth order information. We propose a direct backpropagation-based approach, Smoothed-Autodiff, based on a carefully smoothed version of the original non-differentiable MDP. Our method enjoys low variance at the price of introducing bias, and we theoretically and empirically show that this trade-off can be favorable.
Machine Learning for Health symposium 2022 -- Extended Abstract track
Nov 28, 2022Abstract:A collection of the extended abstracts that were presented at the 2nd Machine Learning for Health symposium (ML4H 2022), which was held both virtually and in person on November 28, 2022, in New Orleans, Louisiana, USA. Machine Learning for Health (ML4H) is a longstanding venue for research into machine learning for health, including both theoretical works and applied works. ML4H 2022 featured two submission tracks: a proceedings track, which encompassed full-length submissions of technically mature and rigorous work, and an extended abstract track, which would accept less mature, but innovative research for discussion. All the manuscripts submitted to ML4H Symposium underwent a double-blind peer-review process. Extended abstracts included in this collection describe innovative machine learning research focused on relevant problems in health and biomedicine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge