Adarsh Subbaswamy
Knowledge-based anomaly detection for identifying network-induced shape artifacts
Nov 06, 2025Abstract:Synthetic data provides a promising approach to address data scarcity for training machine learning models; however, adoption without proper quality assessments may introduce artifacts, distortions, and unrealistic features that compromise model performance and clinical utility. This work introduces a novel knowledge-based anomaly detection method for detecting network-induced shape artifacts in synthetic images. The introduced method utilizes a two-stage framework comprising (i) a novel feature extractor that constructs a specialized feature space by analyzing the per-image distribution of angle gradients along anatomical boundaries, and (ii) an isolation forest-based anomaly detector. We demonstrate the effectiveness of the method for identifying network-induced shape artifacts in two synthetic mammography datasets from models trained on CSAW-M and VinDr-Mammo patient datasets respectively. Quantitative evaluation shows that the method successfully concentrates artifacts in the most anomalous partition (1st percentile), with AUC values of 0.97 (CSAW-syn) and 0.91 (VMLO-syn). In addition, a reader study involving three imaging scientists confirmed that images identified by the method as containing network-induced shape artifacts were also flagged by human readers with mean agreement rates of 66% (CSAW-syn) and 68% (VMLO-syn) for the most anomalous partition, approximately 1.5-2 times higher than the least anomalous partition. Kendall-Tau correlations between algorithmic and human rankings were 0.45 and 0.43 for the two datasets, indicating reasonable agreement despite the challenging nature of subtle artifact detection. This method is a step forward in the responsible use of synthetic data, as it allows developers to evaluate synthetic images for known anatomic constraints and pinpoint and address specific issues to improve the overall quality of a synthetic dataset.
Detecting Dataset Bias in Medical AI: A Generalized and Modality-Agnostic Auditing Framework
Mar 13, 2025Abstract:Data-driven AI is establishing itself at the center of evidence-based medicine. However, reports of shortcomings and unexpected behavior are growing due to AI's reliance on association-based learning. A major reason for this behavior: latent bias in machine learning datasets can be amplified during training and/or hidden during testing. We present a data modality-agnostic auditing framework for generating targeted hypotheses about sources of bias which we refer to as Generalized Attribute Utility and Detectability-Induced bias Testing (G-AUDIT) for datasets. Our method examines the relationship between task-level annotations and data properties including protected attributes (e.g., race, age, sex) and environment and acquisition characteristics (e.g., clinical site, imaging protocols). G-AUDIT automatically quantifies the extent to which the observed data attributes may enable shortcut learning, or in the case of testing data, hide predictions made based on spurious associations. We demonstrate the broad applicability and value of our method by analyzing large-scale medical datasets for three distinct modalities and learning tasks: skin lesion classification in images, stigmatizing language classification in Electronic Health Records (EHR), and mortality prediction for ICU tabular data. In each setting, G-AUDIT successfully identifies subtle biases commonly overlooked by traditional qualitative methods that focus primarily on social and ethical objectives, underscoring its practical value in exposing dataset-level risks and supporting the downstream development of reliable AI systems. Our method paves the way for achieving deeper understanding of machine learning datasets throughout the AI development life-cycle from initial prototyping all the way to regulation, and creates opportunities to reduce model bias, enabling safer and more trustworthy AI systems.
Scorecards for Synthetic Medical Data Evaluation and Reporting
Jun 17, 2024
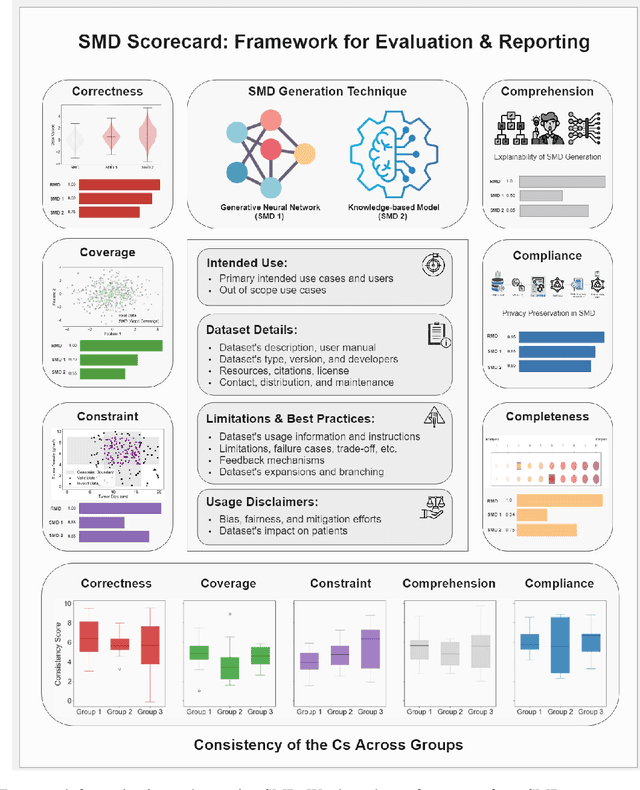
Abstract:The growing utilization of synthetic medical data (SMD) in training and testing AI-driven tools in healthcare necessitates a systematic framework for assessing SMD quality. The current lack of a standardized methodology to evaluate SMD, particularly in terms of its applicability in various medical scenarios, is a significant hindrance to its broader acceptance and utilization in healthcare applications. Here, we outline an evaluation framework designed to meet the unique requirements of medical applications, and introduce the concept of SMD scorecards, which can serve as comprehensive reports that accompany artificially generated datasets. This can help standardize evaluation and enable SMD developers to assess and further enhance the quality of SMDs by identifying areas in need of attention and ensuring that the synthetic data more accurately approximate patient data.
A hierarchical decomposition for explaining ML performance discrepancies
Feb 22, 2024Abstract:Machine learning (ML) algorithms can often differ in performance across domains. Understanding $\textit{why}$ their performance differs is crucial for determining what types of interventions (e.g., algorithmic or operational) are most effective at closing the performance gaps. Existing methods focus on $\textit{aggregate decompositions}$ of the total performance gap into the impact of a shift in the distribution of features $p(X)$ versus the impact of a shift in the conditional distribution of the outcome $p(Y|X)$; however, such coarse explanations offer only a few options for how one can close the performance gap. $\textit{Detailed variable-level decompositions}$ that quantify the importance of each variable to each term in the aggregate decomposition can provide a much deeper understanding and suggest much more targeted interventions. However, existing methods assume knowledge of the full causal graph or make strong parametric assumptions. We introduce a nonparametric hierarchical framework that provides both aggregate and detailed decompositions for explaining why the performance of an ML algorithm differs across domains, without requiring causal knowledge. We derive debiased, computationally-efficient estimators, and statistical inference procedures for asymptotically valid confidence intervals.
Towards a Post-Market Monitoring Framework for Machine Learning-based Medical Devices: A case study
Nov 20, 2023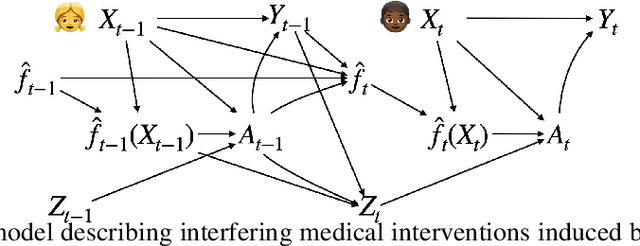
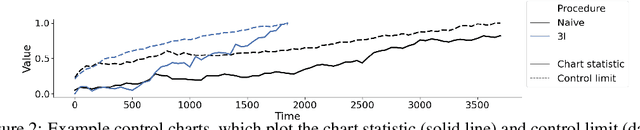
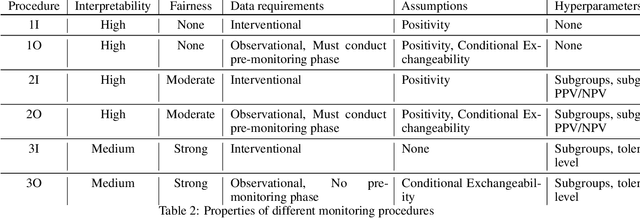
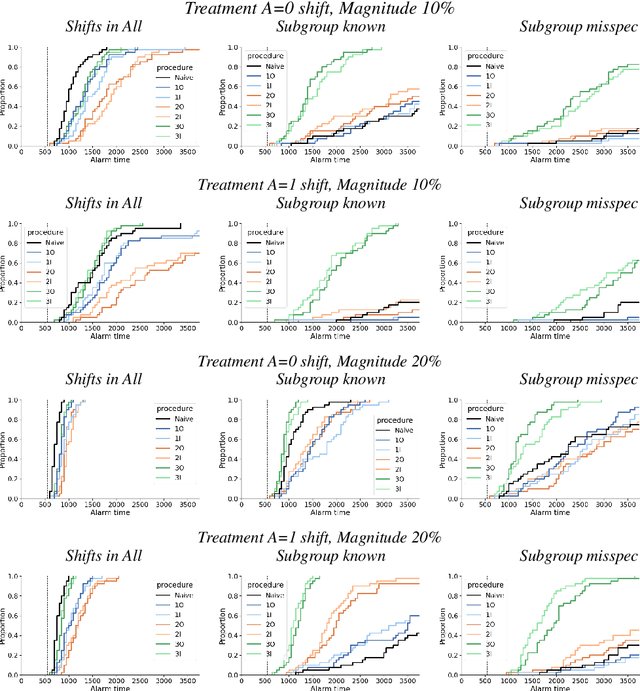
Abstract:After a machine learning (ML)-based system is deployed in clinical practice, performance monitoring is important to ensure the safety and effectiveness of the algorithm over time. The goal of this work is to highlight the complexity of designing a monitoring strategy and the need for a systematic framework that compares the multitude of monitoring options. One of the main decisions is choosing between using real-world (observational) versus interventional data. Although the former is the most convenient source of monitoring data, it exhibits well-known biases, such as confounding, selection, and missingness. In fact, when the ML algorithm interacts with its environment, the algorithm itself may be a primary source of bias. On the other hand, a carefully designed interventional study that randomizes individuals can explicitly eliminate such biases, but the ethics, feasibility, and cost of such an approach must be carefully considered. Beyond the decision of the data source, monitoring strategies vary in the performance criteria they track, the interpretability of the test statistics, the strength of their assumptions, and their speed at detecting performance decay. As a first step towards developing a framework that compares the various monitoring options, we consider a case study of an ML-based risk prediction algorithm for postoperative nausea and vomiting (PONV). Bringing together tools from causal inference and statistical process control, we walk through the basic steps of defining candidate monitoring criteria, describing potential sources of bias and the causal model, and specifying and comparing candidate monitoring procedures. We hypothesize that these steps can be applied more generally, as causal inference can address other sources of biases as well.
Machine Learning for Health symposium 2022 -- Extended Abstract track
Nov 28, 2022Abstract:A collection of the extended abstracts that were presented at the 2nd Machine Learning for Health symposium (ML4H 2022), which was held both virtually and in person on November 28, 2022, in New Orleans, Louisiana, USA. Machine Learning for Health (ML4H) is a longstanding venue for research into machine learning for health, including both theoretical works and applied works. ML4H 2022 featured two submission tracks: a proceedings track, which encompassed full-length submissions of technically mature and rigorous work, and an extended abstract track, which would accept less mature, but innovative research for discussion. All the manuscripts submitted to ML4H Symposium underwent a double-blind peer-review process. Extended abstracts included in this collection describe innovative machine learning research focused on relevant problems in health and biomedicine.
Evaluating Model Robustness to Dataset Shift
Oct 28, 2020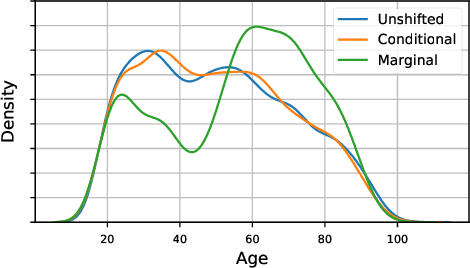



Abstract:As the use of machine learning in safety-critical domains becomes widespread, the importance of evaluating their safety has increased. An important aspect of this is evaluating how robust a model is to changes in setting or population, which typically requires applying the model to multiple, independent datasets. Since the cost of collecting such datasets is often prohibitive, in this paper, we propose a framework for evaluating this type of robustness using a single, fixed evaluation dataset. We use the original evaluation data to define an uncertainty set of possible evaluation distributions and estimate the algorithm's performance on the "worst-case" distribution within this set. Specifically, we consider distribution shifts defined by conditional distributions, allowing some distributions to shift while keeping other portions of the data distribution fixed. This results in finer-grained control over the considered shifts and more plausible worst-case distributions than previous approaches based on covariate shifts. To address the challenges associated with estimation in complex, high-dimensional distributions, we derive a "debiased" estimator which maintains $\sqrt{N}$-consistency even when machine learning methods with slower convergence rates are used to estimate the nuisance parameters. In experiments on a real medical risk prediction task, we show that this estimator can be used to evaluate robustness and accounts for realistic shifts that cannot be expressed as covariate shift. The proposed framework provides a means for practitioners to proactively evaluate the safety of their models using a single validation dataset.
I-SPEC: An End-to-End Framework for Learning Transportable, Shift-Stable Models
Feb 20, 2020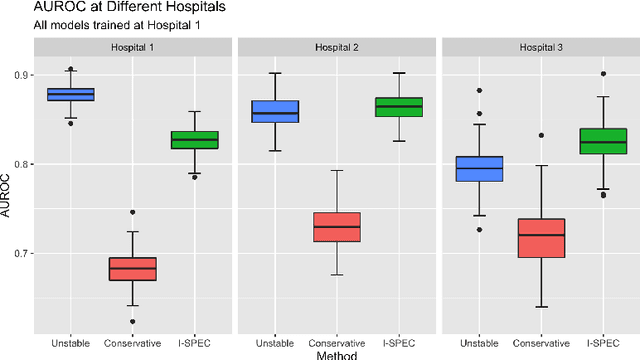
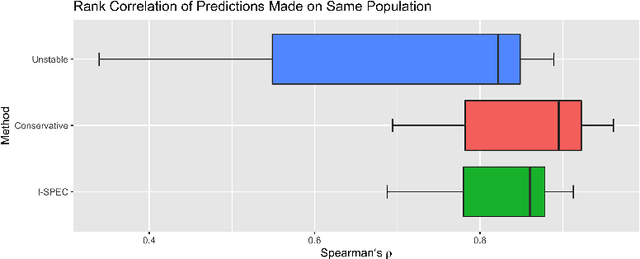
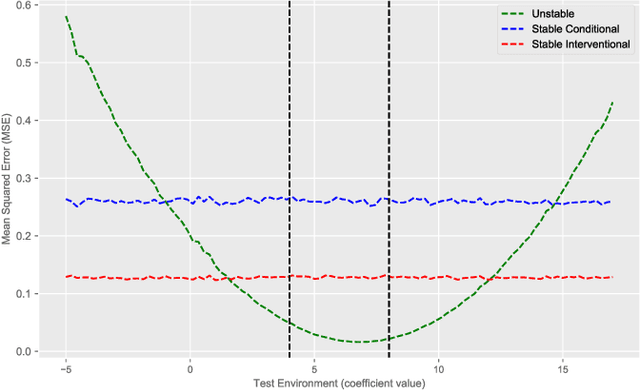
Abstract:Shifts in environment between development and deployment cause classical supervised learning to produce models that fail to generalize well to new target distributions. Recently, many solutions which find invariant predictive distributions have been developed. Among these, graph-based approaches do not require data from the target environment and can capture more stable information than alternative methods which find stable feature sets. However, these approaches assume that the data generating process is known in the form of a full causal graph, which is generally not the case. In this paper, we propose I-SPEC, an end-to-end framework that addresses this shortcoming by using data to learn a partial ancestral graph (PAG). Using the PAG we develop an algorithm that determines an interventional distribution that is stable to the declared shifts; this subsumes existing approaches which find stable feature sets that are less accurate. We apply I-SPEC to a mortality prediction problem to show it can learn a model that is robust to shifts without needing upfront knowledge of the full causal DAG.
The Hierarchy of Stable Distributions and Operators to Trade Off Stability and Performance
May 28, 2019



Abstract:Recent work addressing model reliability and generalization has resulted in a variety of methods that seek to proactively address differences between the training and unknown target environments. While most methods achieve this by finding distributions that will be invariant across environments, we will show they do not necessarily find the same distributions which has implications for performance. In this paper we unify existing work on prediction using stable distributions by relating environmental shifts to edges in the graph underlying a prediction problem, and characterize stable distributions as those which effectively remove these edges. We then quantify the effect of edge deletion on performance in the linear case and corroborate the findings in a simulated and real data experiment.
Tutorial: Safe and Reliable Machine Learning
Apr 15, 2019
Abstract:This document serves as a brief overview of the "Safe and Reliable Machine Learning" tutorial given at the 2019 ACM Conference on Fairness, Accountability, and Transparency (FAT* 2019). The talk slides can be found here: https://bit.ly/2Gfsukp, while a video of the talk is available here: https://youtu.be/FGLOCkC4KmE, and a complete list of references for the tutorial here: https://bit.ly/2GdLPme.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge