Ayah Zirikly
Detecting Dataset Bias in Medical AI: A Generalized and Modality-Agnostic Auditing Framework
Mar 13, 2025Abstract:Data-driven AI is establishing itself at the center of evidence-based medicine. However, reports of shortcomings and unexpected behavior are growing due to AI's reliance on association-based learning. A major reason for this behavior: latent bias in machine learning datasets can be amplified during training and/or hidden during testing. We present a data modality-agnostic auditing framework for generating targeted hypotheses about sources of bias which we refer to as Generalized Attribute Utility and Detectability-Induced bias Testing (G-AUDIT) for datasets. Our method examines the relationship between task-level annotations and data properties including protected attributes (e.g., race, age, sex) and environment and acquisition characteristics (e.g., clinical site, imaging protocols). G-AUDIT automatically quantifies the extent to which the observed data attributes may enable shortcut learning, or in the case of testing data, hide predictions made based on spurious associations. We demonstrate the broad applicability and value of our method by analyzing large-scale medical datasets for three distinct modalities and learning tasks: skin lesion classification in images, stigmatizing language classification in Electronic Health Records (EHR), and mortality prediction for ICU tabular data. In each setting, G-AUDIT successfully identifies subtle biases commonly overlooked by traditional qualitative methods that focus primarily on social and ethical objectives, underscoring its practical value in exposing dataset-level risks and supporting the downstream development of reliable AI systems. Our method paves the way for achieving deeper understanding of machine learning datasets throughout the AI development life-cycle from initial prototyping all the way to regulation, and creates opportunities to reduce model bias, enabling safer and more trustworthy AI systems.
Are Clinical T5 Models Better for Clinical Text?
Dec 08, 2024



Abstract:Large language models with a transformer-based encoder/decoder architecture, such as T5, have become standard platforms for supervised tasks. To bring these technologies to the clinical domain, recent work has trained new or adapted existing models to clinical data. However, the evaluation of these clinical T5 models and comparison to other models has been limited. Are the clinical T5 models better choices than FLAN-tuned generic T5 models? Do they generalize better to new clinical domains that differ from the training sets? We comprehensively evaluate these models across several clinical tasks and domains. We find that clinical T5 models provide marginal improvements over existing models, and perform worse when evaluated on different domains. Our results inform future choices in developing clinical LLMs.
Improving the Generalizability of Depression Detection by Leveraging Clinical Questionnaires
Apr 21, 2022



Abstract:Automated methods have been widely used to identify and analyze mental health conditions (e.g., depression) from various sources of information, including social media. Yet, deployment of such models in real-world healthcare applications faces challenges including poor out-of-domain generalization and lack of trust in black box models. In this work, we propose approaches for depression detection that are constrained to different degrees by the presence of symptoms described in PHQ9, a questionnaire used by clinicians in the depression screening process. In dataset-transfer experiments on three social media datasets, we find that grounding the model in PHQ9's symptoms substantially improves its ability to generalize to out-of-distribution data compared to a standard BERT-based approach. Furthermore, this approach can still perform competitively on in-domain data. These results and our qualitative analyses suggest that grounding model predictions in clinically-relevant symptoms can improve generalizability while producing a model that is easier to inspect.
Sparse encoding for more-interpretable feature-selecting representations in probabilistic matrix factorization
Dec 29, 2020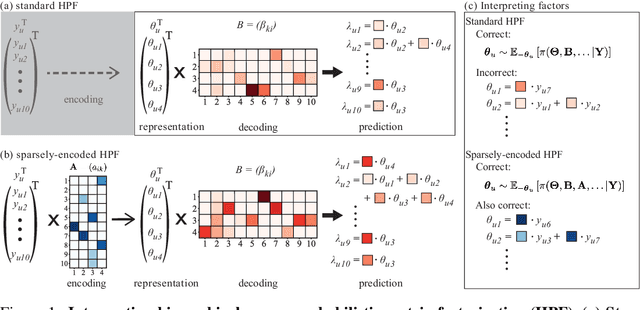

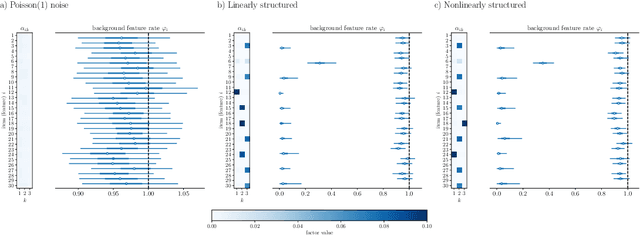

Abstract:Dimensionality reduction methods for count data are critical to a wide range of applications in medical informatics and other fields where model interpretability is paramount. For such data, hierarchical Poisson matrix factorization (HPF) and other sparse probabilistic non-negative matrix factorization (NMF) methods are considered to be interpretable generative models. They consist of sparse transformations for decoding their learned representations into predictions. However, sparsity in representation decoding does not necessarily imply sparsity in the encoding of representations from the original data features. HPF is often incorrectly interpreted in the literature as if it possesses encoder sparsity. The distinction between decoder sparsity and encoder sparsity is subtle but important. Due to the lack of encoder sparsity, HPF does not possess the column-clustering property of classical NMF -- the factor loading matrix does not sufficiently define how each factor is formed from the original features. We address this deficiency by self-consistently enforcing encoder sparsity, using a generalized additive model (GAM), thereby allowing one to relate each representation coordinate to a subset of the original data features. In doing so, the method also gains the ability to perform feature selection. We demonstrate our method on simulated data and give an example of how encoder sparsity is of practical use in a concrete application of representing inpatient comorbidities in Medicare patients.
Classifying the reported ability in clinical mobility descriptions
Jun 07, 2019



Abstract:Assessing how individuals perform different activities is key information for modeling health states of individuals and populations. Descriptions of activity performance in clinical free text are complex, including syntactic negation and similarities to textual entailment tasks. We explore a variety of methods for the novel task of classifying four types of assertions about activity performance: Able, Unable, Unclear, and None (no information). We find that ensembling an SVM trained with lexical features and a CNN achieves 77.9% macro F1 score on our task, and yields nearly 80% recall on the rare Unclear and Unable samples. Finally, we highlight several challenges in classifying performance assertions, including capturing information about sources of assistance, incorporating syntactic structure and negation scope, and handling new modalities at test time. Our findings establish a strong baseline for this novel task, and identify intriguing areas for further research.
RSDD-Time: Temporal Annotation of Self-Reported Mental Health Diagnoses
Jun 20, 2018



Abstract:Self-reported diagnosis statements have been widely employed in studying language related to mental health in social media. However, existing research has largely ignored the temporality of mental health diagnoses. In this work, we introduce RSDD-Time: a new dataset of 598 manually annotated self-reported depression diagnosis posts from Reddit that include temporal information about the diagnosis. Annotations include whether a mental health condition is present and how recently the diagnosis happened. Furthermore, we include exact temporal spans that relate to the date of diagnosis. This information is valuable for various computational methods to examine mental health through social media because one's mental health state is not static. We also test several baseline classification and extraction approaches, which suggest that extracting temporal information from self-reported diagnosis statements is challenging.
Embedding Transfer for Low-Resource Medical Named Entity Recognition: A Case Study on Patient Mobility
Jun 07, 2018



Abstract:Functioning is gaining recognition as an important indicator of global health, but remains under-studied in medical natural language processing research. We present the first analysis of automatically extracting descriptions of patient mobility, using a recently-developed dataset of free text electronic health records. We frame the task as a named entity recognition (NER) problem, and investigate the applicability of NER techniques to mobility extraction. As text corpora focused on patient functioning are scarce, we explore domain adaptation of word embeddings for use in a recurrent neural network NER system. We find that embeddings trained on a small in-domain corpus perform nearly as well as those learned from large out-of-domain corpora, and that domain adaptation techniques yield additional improvements in both precision and recall. Our analysis identifies several significant challenges in extracting descriptions of patient mobility, including the length and complexity of annotated entities and high linguistic variability in mobility descriptions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge