Berkman Sahiner
S-SYNTH: Knowledge-Based, Synthetic Generation of Skin Images
Jul 31, 2024Abstract:Development of artificial intelligence (AI) techniques in medical imaging requires access to large-scale and diverse datasets for training and evaluation. In dermatology, obtaining such datasets remains challenging due to significant variations in patient populations, illumination conditions, and acquisition system characteristics. In this work, we propose S-SYNTH, the first knowledge-based, adaptable open-source skin simulation framework to rapidly generate synthetic skin, 3D models and digitally rendered images, using an anatomically inspired multi-layer, multi-component skin and growing lesion model. The skin model allows for controlled variation in skin appearance, such as skin color, presence of hair, lesion shape, and blood fraction among other parameters. We use this framework to study the effect of possible variations on the development and evaluation of AI models for skin lesion segmentation, and show that results obtained using synthetic data follow similar comparative trends as real dermatologic images, while mitigating biases and limitations from existing datasets including small dataset size, lack of diversity, and underrepresentation.
Image registration based automated lesion correspondence pipeline for longitudinal CT data
Apr 25, 2024



Abstract:Patients diagnosed with metastatic breast cancer (mBC) typically undergo several radiographic assessments during their treatment. mBC often involves multiple metastatic lesions in different organs, it is imperative to accurately track and assess these lesions to gain a comprehensive understanding of the disease's response to treatment. Computerized analysis methods that rely on lesion-level tracking have often used manual matching of corresponding lesions, a time-consuming process that is prone to errors. This paper introduces an automated lesion correspondence algorithm designed to precisely track both targets' lesions and non-targets' lesions in longitudinal data. Here we demonstrate the applicability of our algorithm on the anonymized data from two Phase III trials. The dataset contains imaging data of patients for different follow-up timepoints and the radiologist annotations for the patients enrolled in the trials. Target and non-target lesions are annotated by either one or two groups of radiologists. To facilitate accurate tracking, we have developed a registration-assisted lesion correspondence algorithm. The algorithm employs a sequential two-step pipeline: (a) Firstly, an adaptive Hungarian algorithm is used to establish correspondence among lesions within a single volumetric image series which have been annotated by multiple radiologists at a specific timepoint. (b) Secondly, after establishing correspondence and assigning unique names to the lesions, three-dimensional rigid registration is applied to various image series at the same timepoint. Registration is followed by ongoing lesion correspondence based on the adaptive Hungarian algorithm and updating lesion names for accurate tracking. Validation of our automated lesion correspondence algorithm is performed through triaxial plots based on axial, sagittal, and coronal views, confirming its efficacy in matching lesions.
TorchSurv: A Lightweight Package for Deep Survival Analysis
Apr 17, 2024Abstract:TorchSurv is a Python package that serves as a companion tool to perform deep survival modeling within the PyTorch environment. Unlike existing libraries that impose specific parametric forms, TorchSurv enables the use of custom PyTorch-based deep survival models. With its lightweight design, minimal input requirements, full PyTorch backend, and freedom from restrictive survival model parameterizations, TorchSurv facilitates efficient deep survival model implementation and is particularly beneficial for high-dimensional and complex input data scenarios.
Out-of-Distribution Detection and Data Drift Monitoring using Statistical Process Control
Feb 12, 2024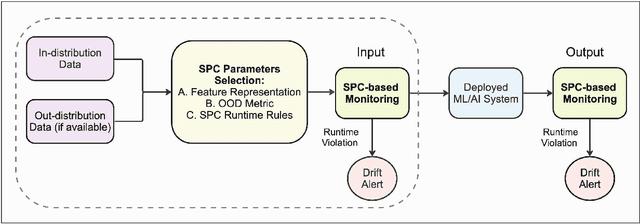

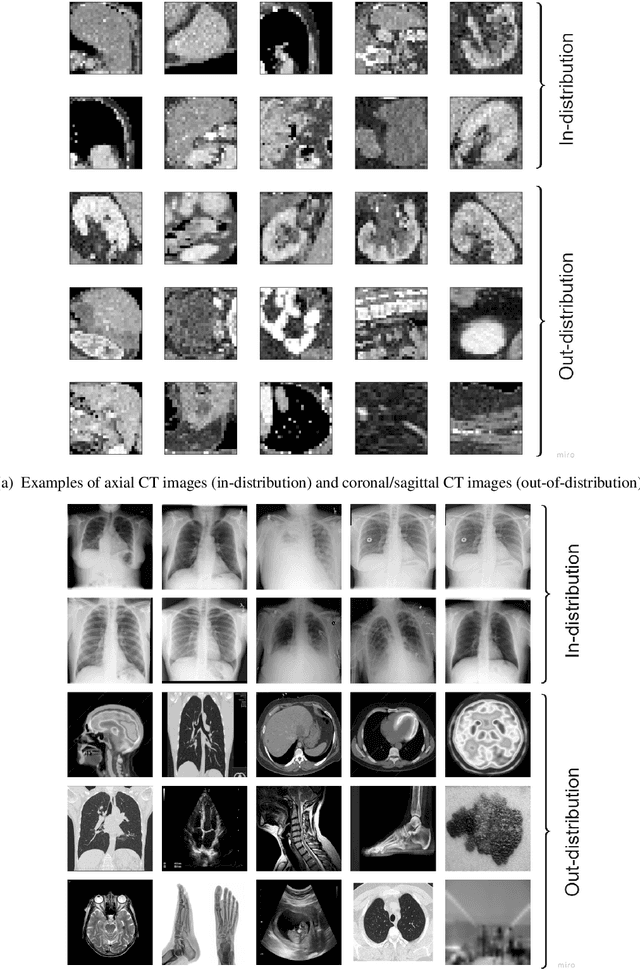
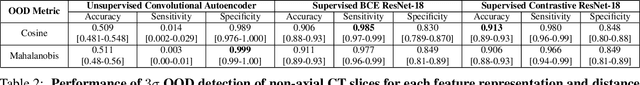
Abstract:Background: Machine learning (ML) methods often fail with data that deviates from their training distribution. This is a significant concern for ML-enabled devices in clinical settings, where data drift may cause unexpected performance that jeopardizes patient safety. Method: We propose a ML-enabled Statistical Process Control (SPC) framework for out-of-distribution (OOD) detection and drift monitoring. SPC is advantageous as it visually and statistically highlights deviations from the expected distribution. To demonstrate the utility of the proposed framework for monitoring data drift in radiological images, we investigated different design choices, including methods for extracting feature representations, drift quantification, and SPC parameter selection. Results: We demonstrate the effectiveness of our framework for two tasks: 1) differentiating axial vs. non-axial computed tomography (CT) images and 2) separating chest x-ray (CXR) from other modalities. For both tasks, we achieved high accuracy in detecting OOD inputs, with 0.913 in CT and 0.995 in CXR, and sensitivity of 0.980 in CT and 0.984 in CXR. Our framework was also adept at monitoring data streams and identifying the time a drift occurred. In a simulation with 100 daily CXR cases, we detected a drift in OOD input percentage from 0-1% to 3-5% within two days, maintaining a low false-positive rate. Through additional experimental results, we demonstrate the framework's data-agnostic nature and independence from the underlying model's structure. Conclusion: We propose a framework for OOD detection and drift monitoring that is agnostic to data, modality, and model. The framework is customizable and can be adapted for specific applications.
Towards a Post-Market Monitoring Framework for Machine Learning-based Medical Devices: A case study
Nov 20, 2023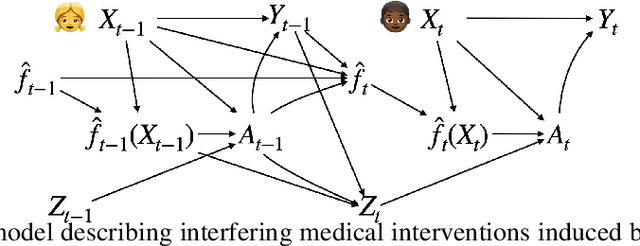
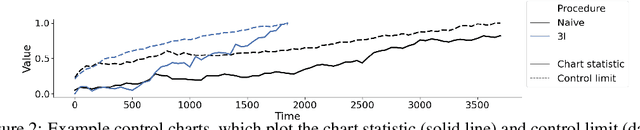
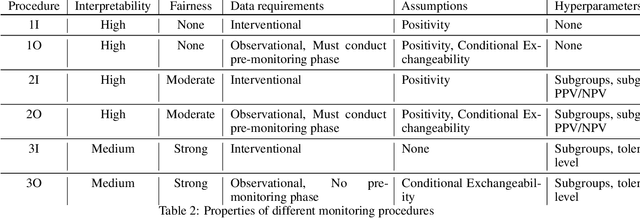
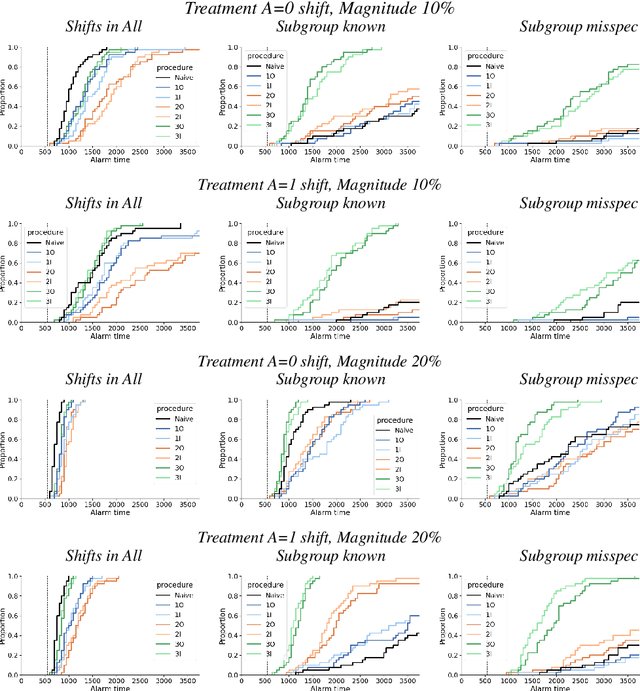
Abstract:After a machine learning (ML)-based system is deployed in clinical practice, performance monitoring is important to ensure the safety and effectiveness of the algorithm over time. The goal of this work is to highlight the complexity of designing a monitoring strategy and the need for a systematic framework that compares the multitude of monitoring options. One of the main decisions is choosing between using real-world (observational) versus interventional data. Although the former is the most convenient source of monitoring data, it exhibits well-known biases, such as confounding, selection, and missingness. In fact, when the ML algorithm interacts with its environment, the algorithm itself may be a primary source of bias. On the other hand, a carefully designed interventional study that randomizes individuals can explicitly eliminate such biases, but the ethics, feasibility, and cost of such an approach must be carefully considered. Beyond the decision of the data source, monitoring strategies vary in the performance criteria they track, the interpretability of the test statistics, the strength of their assumptions, and their speed at detecting performance decay. As a first step towards developing a framework that compares the various monitoring options, we consider a case study of an ML-based risk prediction algorithm for postoperative nausea and vomiting (PONV). Bringing together tools from causal inference and statistical process control, we walk through the basic steps of defining candidate monitoring criteria, describing potential sources of bias and the causal model, and specifying and comparing candidate monitoring procedures. We hypothesize that these steps can be applied more generally, as causal inference can address other sources of biases as well.
Knowledge-based in silico models and dataset for the comparative evaluation of mammography AI for a range of breast characteristics, lesion conspicuities and doses
Oct 27, 2023



Abstract:To generate evidence regarding the safety and efficacy of artificial intelligence (AI) enabled medical devices, AI models need to be evaluated on a diverse population of patient cases, some of which may not be readily available. We propose an evaluation approach for testing medical imaging AI models that relies on in silico imaging pipelines in which stochastic digital models of human anatomy (in object space) with and without pathology are imaged using a digital replica imaging acquisition system to generate realistic synthetic image datasets. Here, we release M-SYNTH, a dataset of cohorts with four breast fibroglandular density distributions imaged at different exposure levels using Monte Carlo x-ray simulations with the publicly available Virtual Imaging Clinical Trial for Regulatory Evaluation (VICTRE) toolkit. We utilize the synthetic dataset to analyze AI model performance and find that model performance decreases with increasing breast density and increases with higher mass density, as expected. As exposure levels decrease, AI model performance drops with the highest performance achieved at exposure levels lower than the nominal recommended dose for the breast type.
Is this model reliable for everyone? Testing for strong calibration
Jul 28, 2023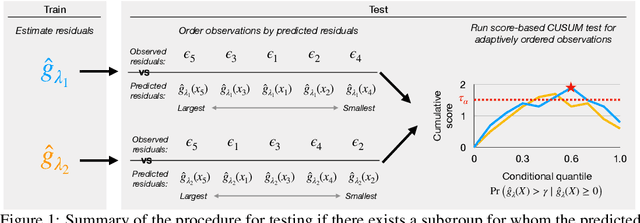

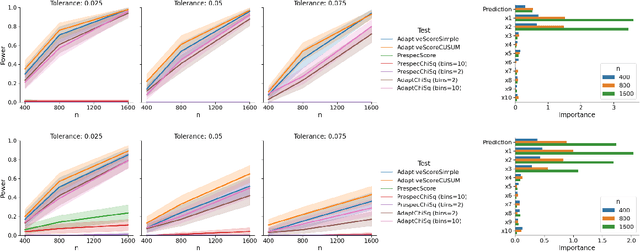
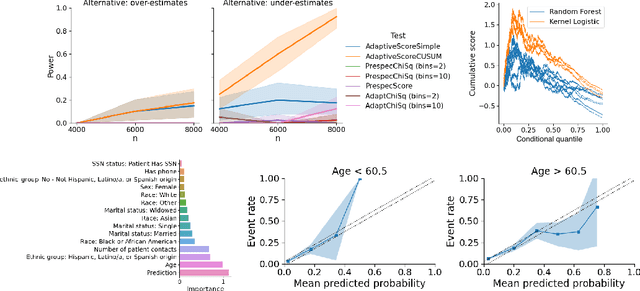
Abstract:In a well-calibrated risk prediction model, the average predicted probability is close to the true event rate for any given subgroup. Such models are reliable across heterogeneous populations and satisfy strong notions of algorithmic fairness. However, the task of auditing a model for strong calibration is well-known to be difficult -- particularly for machine learning (ML) algorithms -- due to the sheer number of potential subgroups. As such, common practice is to only assess calibration with respect to a few predefined subgroups. Recent developments in goodness-of-fit testing offer potential solutions but are not designed for settings with weak signal or where the poorly calibrated subgroup is small, as they either overly subdivide the data or fail to divide the data at all. We introduce a new testing procedure based on the following insight: if we can reorder observations by their expected residuals, there should be a change in the association between the predicted and observed residuals along this sequence if a poorly calibrated subgroup exists. This lets us reframe the problem of calibration testing into one of changepoint detection, for which powerful methods already exist. We begin with introducing a sample-splitting procedure where a portion of the data is used to train a suite of candidate models for predicting the residual, and the remaining data are used to perform a score-based cumulative sum (CUSUM) test. To further improve power, we then extend this adaptive CUSUM test to incorporate cross-validation, while maintaining Type I error control under minimal assumptions. Compared to existing methods, the proposed procedure consistently achieved higher power in simulation studies and more than doubled the power when auditing a mortality risk prediction model.
Monitoring machine learning (ML)-based risk prediction algorithms in the presence of confounding medical interventions
Nov 17, 2022



Abstract:Monitoring the performance of machine learning (ML)-based risk prediction models in healthcare is complicated by the issue of confounding medical interventions (CMI): when an algorithm predicts a patient to be at high risk for an adverse event, clinicians are more likely to administer prophylactic treatment and alter the very target that the algorithm aims to predict. Ignoring CMI by monitoring only the untreated patients--whose outcomes remain unaltered--can inflate false alarm rates, because the evolution of both the model and clinician-ML interactions can induce complex dependencies in the data that violate standard assumptions. A more sophisticated approach is to explicitly account for CMI by modeling treatment propensities, but its time-varying nature makes accurate estimation difficult. Given the many sources of complexity in the data, it is important to determine situations in which a simple procedure that ignores CMI provides valid inference. Here we describe the special case of monitoring model calibration, under either the assumption of conditional exchangeability or time-constant selection bias. We introduce a new score-based cumulative sum (CUSUM) chart for monitoring in a frequentist framework and review an alternative approach using Bayesian inference. Through simulations, we investigate the benefits of combining model updating with monitoring and study when over-trust in a prediction model does (or does not) delay detection. Finally, we simulate monitoring an ML-based postoperative nausea and vomiting risk calculator during the COVID-19 pandemic.
Sequential algorithmic modification with test data reuse
Mar 21, 2022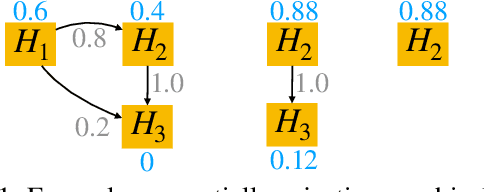
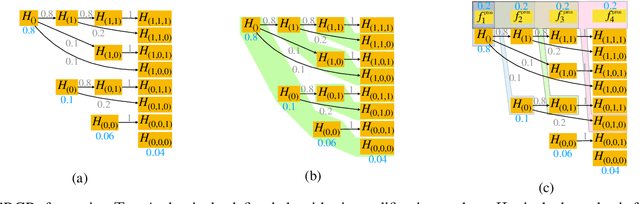
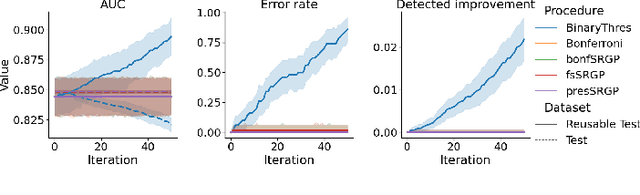
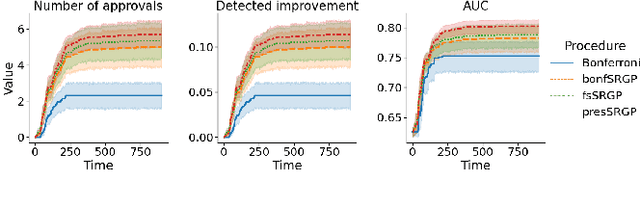
Abstract:After initial release of a machine learning algorithm, the model can be fine-tuned by retraining on subsequently gathered data, adding newly discovered features, or more. Each modification introduces a risk of deteriorating performance and must be validated on a test dataset. It may not always be practical to assemble a new dataset for testing each modification, especially when most modifications are minor or are implemented in rapid succession. Recent works have shown how one can repeatedly test modifications on the same dataset and protect against overfitting by (i) discretizing test results along a grid and (ii) applying a Bonferroni correction to adjust for the total number of modifications considered by an adaptive developer. However, the standard Bonferroni correction is overly conservative when most modifications are beneficial and/or highly correlated. This work investigates more powerful approaches using alpha-recycling and sequentially-rejective graphical procedures (SRGPs). We introduce novel extensions that account for correlation between adaptively chosen algorithmic modifications. In empirical analyses, the SRGPs control the error rate of approving unacceptable modifications and approve a substantially higher number of beneficial modifications than previous approaches.
Bayesian logistic regression for online recalibration and revision of risk prediction models with performance guarantees
Oct 13, 2021



Abstract:After deploying a clinical prediction model, subsequently collected data can be used to fine-tune its predictions and adapt to temporal shifts. Because model updating carries risks of over-updating/fitting, we study online methods with performance guarantees. We introduce two procedures for continual recalibration or revision of an underlying prediction model: Bayesian logistic regression (BLR) and a Markov variant that explicitly models distribution shifts (MarBLR). We perform empirical evaluation via simulations and a real-world study predicting COPD risk. We derive "Type I and II" regret bounds, which guarantee the procedures are non-inferior to a static model and competitive with an oracle logistic reviser in terms of the average loss. Both procedures consistently outperformed the static model and other online logistic revision methods. In simulations, the average estimated calibration index (aECI) of the original model was 0.828 (95%CI 0.818-0.938). Online recalibration using BLR and MarBLR improved the aECI, attaining 0.265 (95%CI 0.230-0.300) and 0.241 (95%CI 0.216-0.266), respectively. When performing more extensive logistic model revisions, BLR and MarBLR increased the average AUC (aAUC) from 0.767 (95%CI 0.765-0.769) to 0.800 (95%CI 0.798-0.802) and 0.799 (95%CI 0.797-0.801), respectively, in stationary settings and protected against substantial model decay. In the COPD study, BLR and MarBLR dynamically combined the original model with a continually-refitted gradient boosted tree to achieve aAUCs of 0.924 (95%CI 0.913-0.935) and 0.925 (95%CI 0.914-0.935), compared to the static model's aAUC of 0.904 (95%CI 0.892-0.916). Despite its simplicity, BLR is highly competitive with MarBLR. MarBLR outperforms BLR when its prior better reflects the data. BLR and MarBLR can improve the transportability of clinical prediction models and maintain their performance over time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge