Alexej Gossmann
DomainLab: A modular Python package for domain generalization in deep learning
Mar 21, 2024
Abstract:Poor generalization performance caused by distribution shifts in unseen domains often hinders the trustworthy deployment of deep neural networks. Many domain generalization techniques address this problem by adding a domain invariant regularization loss terms during training. However, there is a lack of modular software that allows users to combine the advantages of different methods with minimal effort for reproducibility. DomainLab is a modular Python package for training user specified neural networks with composable regularization loss terms. Its decoupled design allows the separation of neural networks from regularization loss construction. Hierarchical combinations of neural networks, different domain generalization methods, and associated hyperparameters, can all be specified together with other experimental setup in a single configuration file. Hierarchical combinations of neural networks, different domain generalization methods, and associated hyperparameters, can all be specified together with other experimental setup in a single configuration file. In addition, DomainLab offers powerful benchmarking functionality to evaluate the generalization performance of neural networks in out-of-distribution data. The package supports running the specified benchmark on an HPC cluster or on a standalone machine. The package is well tested with over 95 percent coverage and well documented. From the user perspective, it is closed to modification but open to extension. The package is under the MIT license, and its source code, tutorial and documentation can be found at https://github.com/marrlab/DomainLab.
M-HOF-Opt: Multi-Objective Hierarchical Output Feedback Optimization via Multiplier Induced Loss Landscape Scheduling
Mar 20, 2024



Abstract:When a neural network parameterized loss function consists of many terms, the combinatorial choice of weight multipliers during the optimization process forms a challenging problem. To address this, we proposed a probabilistic graphical model (PGM) for the joint model parameter and multiplier evolution process, with a hypervolume based likelihood that promotes multi-objective descent of each loss term. The corresponding parameter and multiplier estimation as a sequential decision process is then cast into an optimal control problem, where the multi-objective descent goal is dispatched hierarchically into a series of constraint optimization sub-problems. The sub-problem constraint automatically adapts itself according to Pareto dominance and serves as the setpoint for the low level multiplier controller to schedule loss landscapes via output feedback of each loss term. Our method is multiplier-free and operates at the timescale of epochs, thus saves tremendous computational resources compared to full training cycle multiplier tuning. We applied it to domain invariant variational auto-encoding with 6 loss terms on the PACS domain generalization task, and observed robust performance across a range of controller hyperparameters, as well as different multiplier initial conditions, outperforming other multiplier scheduling methods. We offered modular implementation of our method, admitting custom definition of many loss terms for applying our multi-objective hierarchical output feedback training scheme to other deep learning fields.
A hierarchical decomposition for explaining ML performance discrepancies
Feb 22, 2024Abstract:Machine learning (ML) algorithms can often differ in performance across domains. Understanding $\textit{why}$ their performance differs is crucial for determining what types of interventions (e.g., algorithmic or operational) are most effective at closing the performance gaps. Existing methods focus on $\textit{aggregate decompositions}$ of the total performance gap into the impact of a shift in the distribution of features $p(X)$ versus the impact of a shift in the conditional distribution of the outcome $p(Y|X)$; however, such coarse explanations offer only a few options for how one can close the performance gap. $\textit{Detailed variable-level decompositions}$ that quantify the importance of each variable to each term in the aggregate decomposition can provide a much deeper understanding and suggest much more targeted interventions. However, existing methods assume knowledge of the full causal graph or make strong parametric assumptions. We introduce a nonparametric hierarchical framework that provides both aggregate and detailed decompositions for explaining why the performance of an ML algorithm differs across domains, without requiring causal knowledge. We derive debiased, computationally-efficient estimators, and statistical inference procedures for asymptotically valid confidence intervals.
Towards a Post-Market Monitoring Framework for Machine Learning-based Medical Devices: A case study
Nov 20, 2023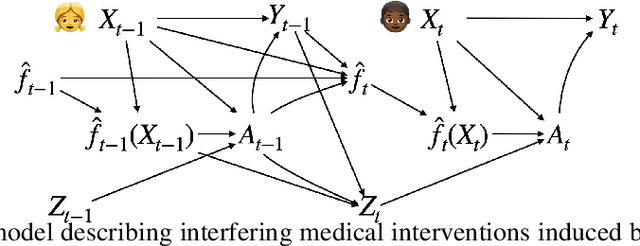
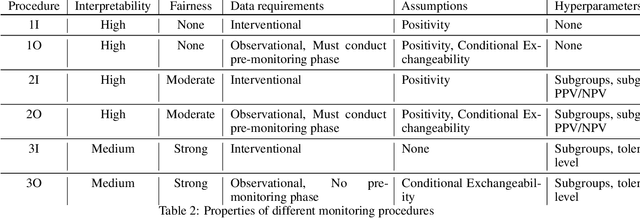
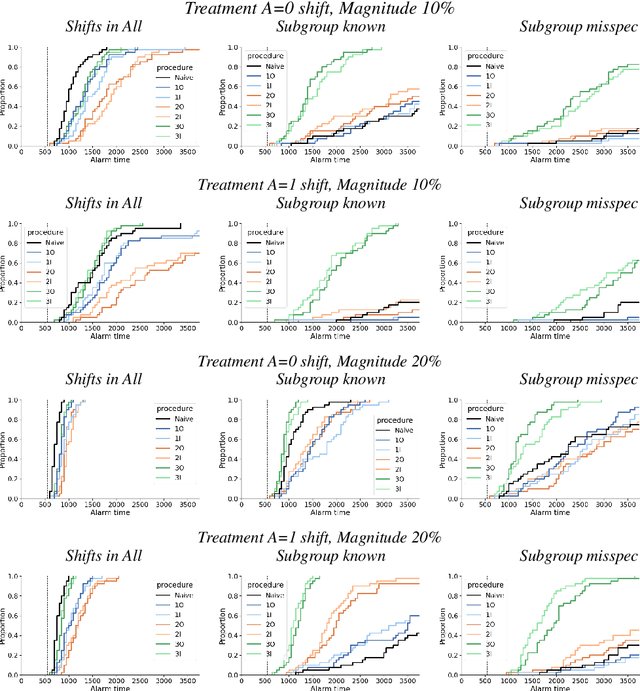
Abstract:After a machine learning (ML)-based system is deployed in clinical practice, performance monitoring is important to ensure the safety and effectiveness of the algorithm over time. The goal of this work is to highlight the complexity of designing a monitoring strategy and the need for a systematic framework that compares the multitude of monitoring options. One of the main decisions is choosing between using real-world (observational) versus interventional data. Although the former is the most convenient source of monitoring data, it exhibits well-known biases, such as confounding, selection, and missingness. In fact, when the ML algorithm interacts with its environment, the algorithm itself may be a primary source of bias. On the other hand, a carefully designed interventional study that randomizes individuals can explicitly eliminate such biases, but the ethics, feasibility, and cost of such an approach must be carefully considered. Beyond the decision of the data source, monitoring strategies vary in the performance criteria they track, the interpretability of the test statistics, the strength of their assumptions, and their speed at detecting performance decay. As a first step towards developing a framework that compares the various monitoring options, we consider a case study of an ML-based risk prediction algorithm for postoperative nausea and vomiting (PONV). Bringing together tools from causal inference and statistical process control, we walk through the basic steps of defining candidate monitoring criteria, describing potential sources of bias and the causal model, and specifying and comparing candidate monitoring procedures. We hypothesize that these steps can be applied more generally, as causal inference can address other sources of biases as well.
Is this model reliable for everyone? Testing for strong calibration
Jul 28, 2023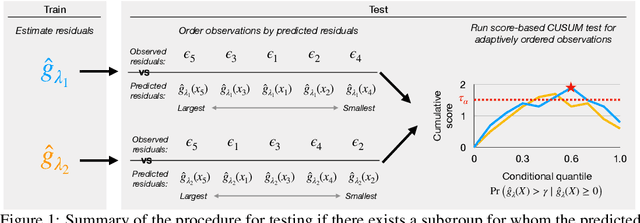

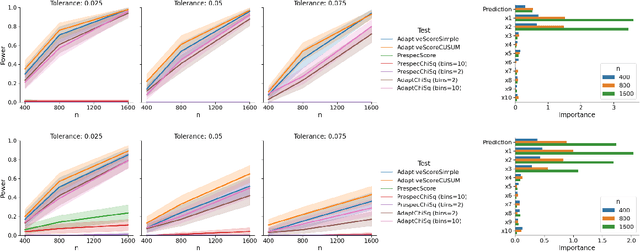
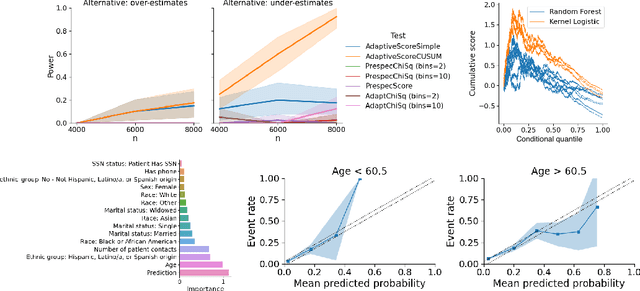
Abstract:In a well-calibrated risk prediction model, the average predicted probability is close to the true event rate for any given subgroup. Such models are reliable across heterogeneous populations and satisfy strong notions of algorithmic fairness. However, the task of auditing a model for strong calibration is well-known to be difficult -- particularly for machine learning (ML) algorithms -- due to the sheer number of potential subgroups. As such, common practice is to only assess calibration with respect to a few predefined subgroups. Recent developments in goodness-of-fit testing offer potential solutions but are not designed for settings with weak signal or where the poorly calibrated subgroup is small, as they either overly subdivide the data or fail to divide the data at all. We introduce a new testing procedure based on the following insight: if we can reorder observations by their expected residuals, there should be a change in the association between the predicted and observed residuals along this sequence if a poorly calibrated subgroup exists. This lets us reframe the problem of calibration testing into one of changepoint detection, for which powerful methods already exist. We begin with introducing a sample-splitting procedure where a portion of the data is used to train a suite of candidate models for predicting the residual, and the remaining data are used to perform a score-based cumulative sum (CUSUM) test. To further improve power, we then extend this adaptive CUSUM test to incorporate cross-validation, while maintaining Type I error control under minimal assumptions. Compared to existing methods, the proposed procedure consistently achieved higher power in simulation studies and more than doubled the power when auditing a mortality risk prediction model.
Monitoring machine learning (ML)-based risk prediction algorithms in the presence of confounding medical interventions
Nov 17, 2022



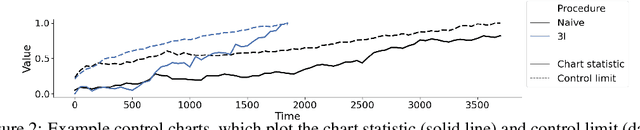
Abstract:Monitoring the performance of machine learning (ML)-based risk prediction models in healthcare is complicated by the issue of confounding medical interventions (CMI): when an algorithm predicts a patient to be at high risk for an adverse event, clinicians are more likely to administer prophylactic treatment and alter the very target that the algorithm aims to predict. Ignoring CMI by monitoring only the untreated patients--whose outcomes remain unaltered--can inflate false alarm rates, because the evolution of both the model and clinician-ML interactions can induce complex dependencies in the data that violate standard assumptions. A more sophisticated approach is to explicitly account for CMI by modeling treatment propensities, but its time-varying nature makes accurate estimation difficult. Given the many sources of complexity in the data, it is important to determine situations in which a simple procedure that ignores CMI provides valid inference. Here we describe the special case of monitoring model calibration, under either the assumption of conditional exchangeability or time-constant selection bias. We introduce a new score-based cumulative sum (CUSUM) chart for monitoring in a frequentist framework and review an alternative approach using Bayesian inference. Through simulations, we investigate the benefits of combining model updating with monitoring and study when over-trust in a prediction model does (or does not) delay detection. Finally, we simulate monitoring an ML-based postoperative nausea and vomiting risk calculator during the COVID-19 pandemic.
Sequential algorithmic modification with test data reuse
Mar 21, 2022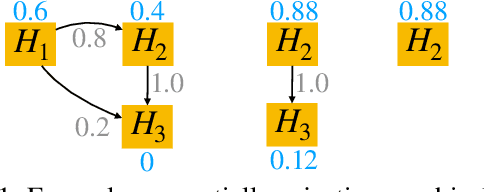
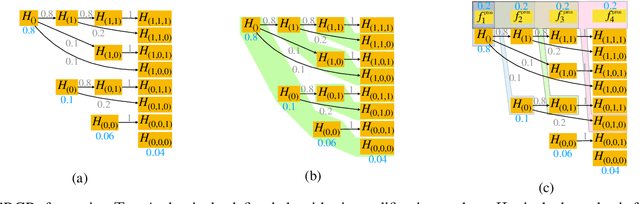
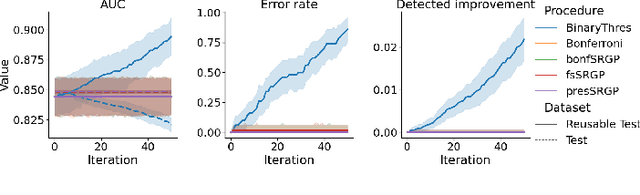
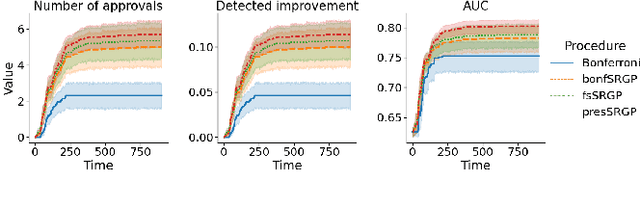
Abstract:After initial release of a machine learning algorithm, the model can be fine-tuned by retraining on subsequently gathered data, adding newly discovered features, or more. Each modification introduces a risk of deteriorating performance and must be validated on a test dataset. It may not always be practical to assemble a new dataset for testing each modification, especially when most modifications are minor or are implemented in rapid succession. Recent works have shown how one can repeatedly test modifications on the same dataset and protect against overfitting by (i) discretizing test results along a grid and (ii) applying a Bonferroni correction to adjust for the total number of modifications considered by an adaptive developer. However, the standard Bonferroni correction is overly conservative when most modifications are beneficial and/or highly correlated. This work investigates more powerful approaches using alpha-recycling and sequentially-rejective graphical procedures (SRGPs). We introduce novel extensions that account for correlation between adaptively chosen algorithmic modifications. In empirical analyses, the SRGPs control the error rate of approving unacceptable modifications and approve a substantially higher number of beneficial modifications than previous approaches.
Bayesian logistic regression for online recalibration and revision of risk prediction models with performance guarantees
Oct 13, 2021



Abstract:After deploying a clinical prediction model, subsequently collected data can be used to fine-tune its predictions and adapt to temporal shifts. Because model updating carries risks of over-updating/fitting, we study online methods with performance guarantees. We introduce two procedures for continual recalibration or revision of an underlying prediction model: Bayesian logistic regression (BLR) and a Markov variant that explicitly models distribution shifts (MarBLR). We perform empirical evaluation via simulations and a real-world study predicting COPD risk. We derive "Type I and II" regret bounds, which guarantee the procedures are non-inferior to a static model and competitive with an oracle logistic reviser in terms of the average loss. Both procedures consistently outperformed the static model and other online logistic revision methods. In simulations, the average estimated calibration index (aECI) of the original model was 0.828 (95%CI 0.818-0.938). Online recalibration using BLR and MarBLR improved the aECI, attaining 0.265 (95%CI 0.230-0.300) and 0.241 (95%CI 0.216-0.266), respectively. When performing more extensive logistic model revisions, BLR and MarBLR increased the average AUC (aAUC) from 0.767 (95%CI 0.765-0.769) to 0.800 (95%CI 0.798-0.802) and 0.799 (95%CI 0.797-0.801), respectively, in stationary settings and protected against substantial model decay. In the COPD study, BLR and MarBLR dynamically combined the original model with a continually-refitted gradient boosted tree to achieve aAUCs of 0.924 (95%CI 0.913-0.935) and 0.925 (95%CI 0.914-0.935), compared to the static model's aAUC of 0.904 (95%CI 0.892-0.916). Despite its simplicity, BLR is highly competitive with MarBLR. MarBLR outperforms BLR when its prior better reflects the data. BLR and MarBLR can improve the transportability of clinical prediction models and maintain their performance over time.
Resampling-based Assessment of Robustness to Distribution Shift for Deep Neural Networks
Jun 07, 2019



Abstract:A novel resampling framework is proposed to evaluate the robustness and generalization capability of deep learning models with respect to distribution shift. We use Auto Encoder Variational Bayes to find a latent representation of the data, on which a Variational Gaussian Mixture Model is applied to deliberately create distribution shift by dividing the dataset into different clusters. Wasserstein distance is used to characterize the extent of distribution shift between the training and the testing data splits. We compare several conventional Convolutional Neural Network (CNN) architectures as well as Bayesian CNN models for image classification on the Fashion-MNIST dataset to assess their robustness under the deliberately created distribution shift.
Multimodal Sparse Classifier for Adolescent Brain Age Prediction
Apr 01, 2019
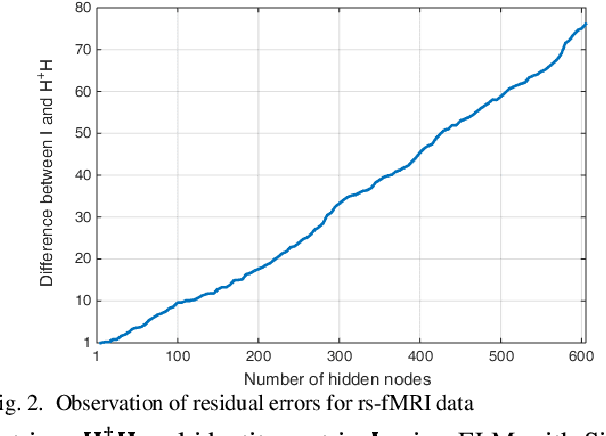
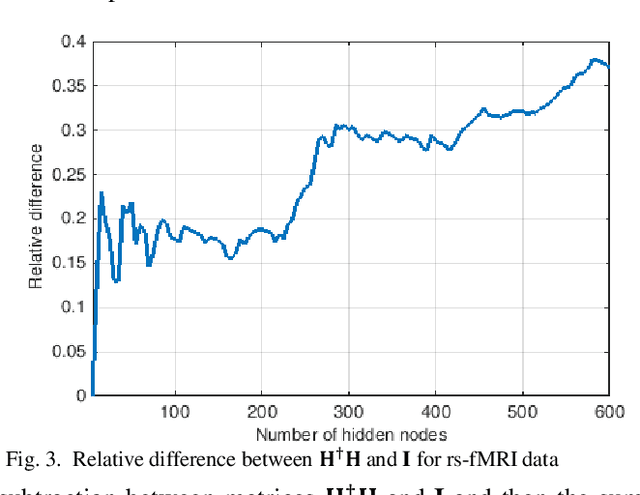
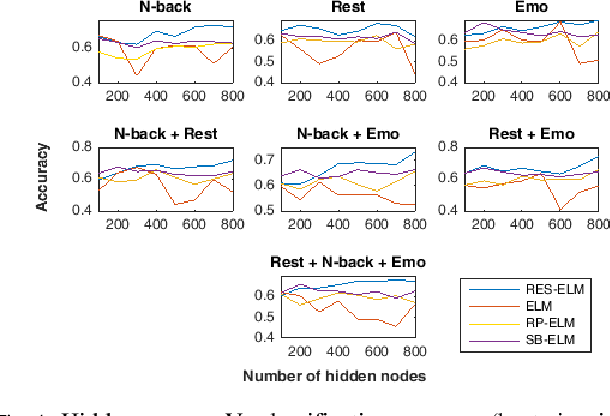
Abstract:The study of healthy brain development helps to better understand the brain transformation and brain connectivity patterns which happen during childhood to adulthood. This study presents a sparse machine learning solution across whole-brain functional connectivity (FC) measures of three sets of data, derived from resting state functional magnetic resonance imaging (rs-fMRI) and task fMRI data, including a working memory n-back task (nb-fMRI) and an emotion identification task (em-fMRI). These multi-modal image data are collected on a sample of adolescents from the Philadelphia Neurodevelopmental Cohort (PNC) for the prediction of brain ages. Due to extremely large variable-to-instance ratio of PNC data, a high dimensional matrix with several irrelevant and highly correlated features is generated and hence a pattern learning approach is necessary to extract significant features. We propose a sparse learner based on the residual errors along the estimation of an inverse problem for the extreme learning machine (ELM) neural network. The purpose of the approach is to overcome the overlearning problem through pruning of several redundant features and their corresponding output weights. The proposed multimodal sparse ELM classifier based on residual errors (RES-ELM) is highly competitive in terms of the classification accuracy compared to its counterparts such as conventional ELM, and sparse Bayesian learning ELM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge