Libo Ren
MaLei at MultiClinSUM: Summarisation of Clinical Documents using Perspective-Aware Iterative Self-Prompting with LLMs
Sep 09, 2025Abstract:Efficient communication between patients and clinicians plays an important role in shared decision-making. However, clinical reports are often lengthy and filled with clinical jargon, making it difficult for domain experts to identify important aspects in the document efficiently. This paper presents the methodology we applied in the MultiClinSUM shared task for summarising clinical case documents. We used an Iterative Self-Prompting technique on large language models (LLMs) by asking LLMs to generate task-specific prompts and refine them via example-based few-shot learning. Furthermore, we used lexical and embedding space metrics, ROUGE and BERT-score, to guide the model fine-tuning with epochs. Our submission using perspective-aware ISP on GPT-4 and GPT-4o achieved ROUGE scores (46.53, 24.68, 30.77) and BERTscores (87.84, 83.25, 85.46) for (P, R, F1) from the official evaluation on 3,396 clinical case reports from various specialties extracted from open journals. The high BERTscore indicates that the model produced semantically equivalent output summaries compared to the references, even though the overlap at the exact lexicon level is lower, as reflected in the lower ROUGE scores. This work sheds some light on how perspective-aware ISP (PA-ISP) can be deployed for clinical report summarisation and support better communication between patients and clinicians.
Generating Synthetic Free-text Medical Records with Low Re-identification Risk using Masked Language Modeling
Sep 17, 2024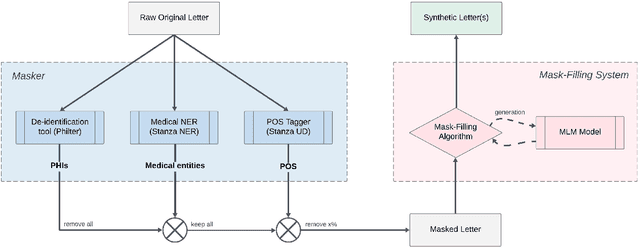

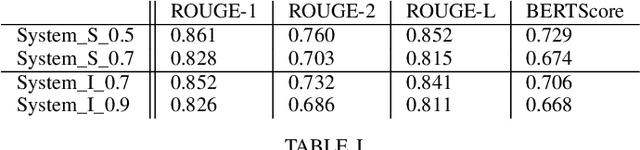
Abstract:In this paper, we present a system that generates synthetic free-text medical records, such as discharge summaries, admission notes and doctor correspondences, using Masked Language Modeling (MLM). Our system is designed to preserve the critical information of the records while introducing significant diversity and minimizing re-identification risk. The system incorporates a de-identification component that uses Philter to mask Protected Health Information (PHI), followed by a Medical Entity Recognition (NER) model to retain key medical information. We explore various masking ratios and mask-filling techniques to balance the trade-off between diversity and fidelity in the synthetic outputs without affecting overall readability. Our results demonstrate that the system can produce high-quality synthetic data with significant diversity while achieving a HIPAA-compliant PHI recall rate of 0.96 and a low re-identification risk of 0.035. Furthermore, downstream evaluations using a NER task reveal that the synthetic data can be effectively used to train models with performance comparable to those trained on real data. The flexibility of the system allows it to be adapted for specific use cases, making it a valuable tool for privacy-preserving data generation in medical research and healthcare applications.
Synthetic4Health: Generating Annotated Synthetic Clinical Letters
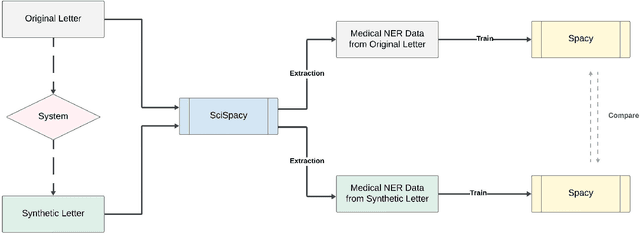
Sep 14, 2024Abstract:Since clinical letters contain sensitive information, clinical-related datasets can not be widely applied in model training, medical research, and teaching. This work aims to generate reliable, various, and de-identified synthetic clinical letters. To achieve this goal, we explored different pre-trained language models (PLMs) for masking and generating text. After that, we worked on Bio\_ClinicalBERT, a high-performing model, and experimented with different masking strategies. Both qualitative and quantitative methods were used for evaluation. Additionally, a downstream task, Named Entity Recognition (NER), was also implemented to assess the usability of these synthetic letters. The results indicate that 1) encoder-only models outperform encoder-decoder models. 2) Among encoder-only models, those trained on general corpora perform comparably to those trained on clinical data when clinical information is preserved. 3) Additionally, preserving clinical entities and document structure better aligns with our objectives than simply fine-tuning the model. 4) Furthermore, different masking strategies can impact the quality of synthetic clinical letters. Masking stopwords has a positive impact, while masking nouns or verbs has a negative effect. 5) For evaluation, BERTScore should be the primary quantitative evaluation metric, with other metrics serving as supplementary references. 6) Contextual information does not significantly impact the models' understanding, so the synthetic clinical letters have the potential to replace the original ones in downstream tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge