Prakash Adekkanattu
Extracting Social Support and Social Isolation Information from Clinical Psychiatry Notes: Comparing a Rule-based NLP System and a Large Language Model
Mar 25, 2024Abstract:Background: Social support (SS) and social isolation (SI) are social determinants of health (SDOH) associated with psychiatric outcomes. In electronic health records (EHRs), individual-level SS/SI is typically documented as narrative clinical notes rather than structured coded data. Natural language processing (NLP) algorithms can automate the otherwise labor-intensive process of data extraction. Data and Methods: Psychiatric encounter notes from Mount Sinai Health System (MSHS, n=300) and Weill Cornell Medicine (WCM, n=225) were annotated and established a gold standard corpus. A rule-based system (RBS) involving lexicons and a large language model (LLM) using FLAN-T5-XL were developed to identify mentions of SS and SI and their subcategories (e.g., social network, instrumental support, and loneliness). Results: For extracting SS/SI, the RBS obtained higher macro-averaged f-scores than the LLM at both MSHS (0.89 vs. 0.65) and WCM (0.85 vs. 0.82). For extracting subcategories, the RBS also outperformed the LLM at both MSHS (0.90 vs. 0.62) and WCM (0.82 vs. 0.81). Discussion and Conclusion: Unexpectedly, the RBS outperformed the LLMs across all metrics. Intensive review demonstrates that this finding is due to the divergent approach taken by the RBS and LLM. The RBS were designed and refined to follow the same specific rules as the gold standard annotations. Conversely, the LLM were more inclusive with categorization and conformed to common English-language understanding. Both approaches offer advantages and are made available open-source for future testing.
SODA: A Natural Language Processing Package to Extract Social Determinants of Health for Cancer Studies
Dec 06, 2022



Abstract:Objective: We aim to develop an open-source natural language processing (NLP) package, SODA (i.e., SOcial DeterminAnts), with pre-trained transformer models to extract social determinants of health (SDoH) for cancer patients, examine the generalizability of SODA to a new disease domain (i.e., opioid use), and evaluate the extraction rate of SDoH using cancer populations. Methods: We identified SDoH categories and attributes and developed an SDoH corpus using clinical notes from a general cancer cohort. We compared four transformer-based NLP models to extract SDoH, examined the generalizability of NLP models to a cohort of patients prescribed with opioids, and explored customization strategies to improve performance. We applied the best NLP model to extract 19 categories of SDoH from the breast (n=7,971), lung (n=11,804), and colorectal cancer (n=6,240) cohorts. Results and Conclusion: We developed a corpus of 629 cancer patients notes with annotations of 13,193 SDoH concepts/attributes from 19 categories of SDoH. The Bidirectional Encoder Representations from Transformers (BERT) model achieved the best strict/lenient F1 scores of 0.9216 and 0.9441 for SDoH concept extraction, 0.9617 and 0.9626 for linking attributes to SDoH concepts. Fine-tuning the NLP models using new annotations from opioid use patients improved the strict/lenient F1 scores from 0.8172/0.8502 to 0.8312/0.8679. The extraction rates among 19 categories of SDoH varied greatly, where 10 SDoH could be extracted from >70% of cancer patients, but 9 SDoH had a low extraction rate (<70% of cancer patients). The SODA package with pre-trained transformer models is publicly available at https://github.com/uf-hobiinformatics-lab/SDoH_SODA.
A Study of Social and Behavioral Determinants of Health in Lung Cancer Patients Using Transformers-based Natural Language Processing Models
Aug 10, 2021



Abstract:Social and behavioral determinants of health (SBDoH) have important roles in shaping people's health. In clinical research studies, especially comparative effectiveness studies, failure to adjust for SBDoH factors will potentially cause confounding issues and misclassification errors in either statistical analyses and machine learning-based models. However, there are limited studies to examine SBDoH factors in clinical outcomes due to the lack of structured SBDoH information in current electronic health record (EHR) systems, while much of the SBDoH information is documented in clinical narratives. Natural language processing (NLP) is thus the key technology to extract such information from unstructured clinical text. However, there is not a mature clinical NLP system focusing on SBDoH. In this study, we examined two state-of-the-art transformer-based NLP models, including BERT and RoBERTa, to extract SBDoH concepts from clinical narratives, applied the best performing model to extract SBDoH concepts on a lung cancer screening patient cohort, and examined the difference of SBDoH information between NLP extracted results and structured EHRs (SBDoH information captured in standard vocabularies such as the International Classification of Diseases codes). The experimental results show that the BERT-based NLP model achieved the best strict/lenient F1-score of 0.8791 and 0.8999, respectively. The comparison between NLP extracted SBDoH information and structured EHRs in the lung cancer patient cohort of 864 patients with 161,933 various types of clinical notes showed that much more detailed information about smoking, education, and employment were only captured in clinical narratives and that it is necessary to use both clinical narratives and structured EHRs to construct a more complete picture of patients' SBDoH factors.
Identification of Predictive Sub-Phenotypes of Acute Kidney Injury using Structured and Unstructured Electronic Health Record Data with Memory Networks
Apr 10, 2019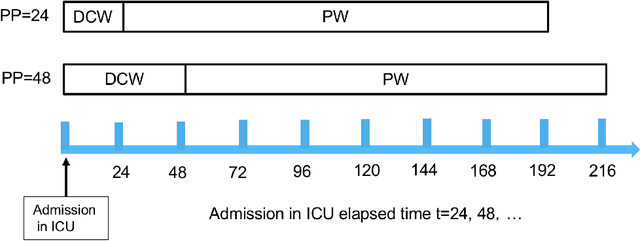
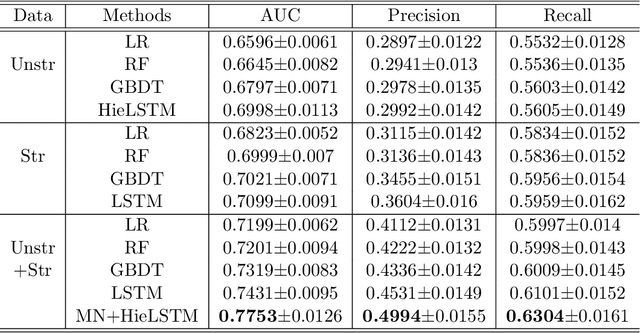
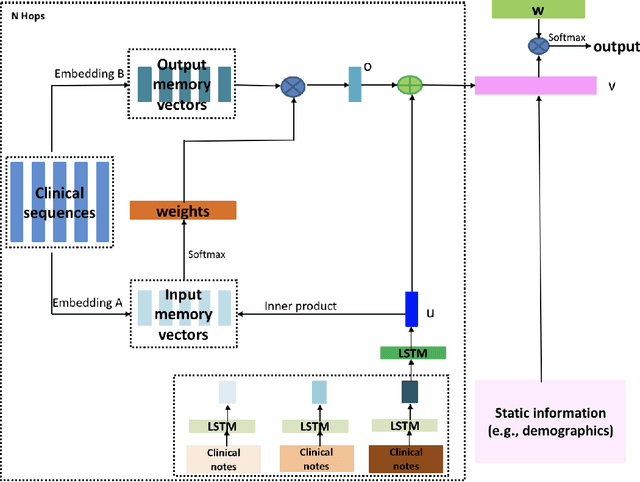
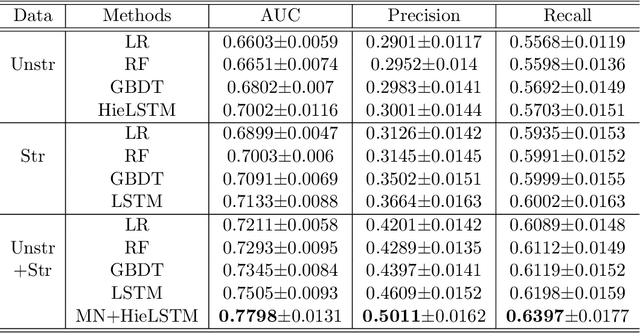
Abstract:Acute Kidney Injury (AKI) is a common clinical syndrome characterized by the rapid loss of kidney excretory function, which aggravates the clinical severity of other diseases in a large number of hospitalized patients. Accurate early prediction of AKI can enable in-time interventions and treatments. However, AKI is highly heterogeneous, thus identification of AKI sub-phenotypes can lead to an improved understanding of the disease pathophysiology and development of more targeted clinical interventions. This study used a memory network-based deep learning approach to discover predictive AKI sub-phenotypes using structured and unstructured electronic health record (EHR) data of patients before AKI diagnosis. We leveraged a real world critical care EHR corpus including 37,486 ICU stays. Our approach identified three distinct sub-phenotypes: sub-phenotype I is with an average age of 63.03$ \pm 17.25 $ years, and is characterized by mild loss of kidney excretory function (Serum Creatinne (SCr) $1.55\pm 0.34$ mg/dL, estimated Glomerular Filtration Rate Test (eGFR) $107.65\pm 54.98$ mL/min/1.73$m^2$). These patients are more likely to develop stage I AKI. Sub-phenotype II is with average age 66.81$ \pm 10.43 $ years, and was characterized by severe loss of kidney excretory function (SCr $1.96\pm 0.49$ mg/dL, eGFR $82.19\pm 55.92$ mL/min/1.73$m^2$). These patients are more likely to develop stage III AKI. Sub-phenotype III is with average age 65.07$ \pm 11.32 $ years, and was characterized moderate loss of kidney excretory function and thus more likely to develop stage II AKI (SCr $1.69\pm 0.32$ mg/dL, eGFR $93.97\pm 56.53$ mL/min/1.73$m^2$). Both SCr and eGFR are significantly different across the three sub-phenotypes with statistical testing plus postdoc analysis, and the conclusion still holds after age adjustment.
Evaluating the Portability of an NLP System for Processing Echocardiograms: A Retrospective, Multi-site Observational Study
Apr 02, 2019

Abstract:While natural language processing (NLP) of unstructured clinical narratives holds the potential for patient care and clinical research, portability of NLP approaches across multiple sites remains a major challenge. This study investigated the portability of an NLP system developed initially at the Department of Veterans Affairs (VA) to extract 27 key cardiac concepts from free-text or semi-structured echocardiograms from three academic medical centers: Weill Cornell Medicine, Mayo Clinic and Northwestern Medicine. While the NLP system showed high precision and recall measurements for four target concepts (aortic valve regurgitation, left atrium size at end systole, mitral valve regurgitation, tricuspid valve regurgitation) across all sites, we found moderate or poor results for the remaining concepts and the NLP system performance varied between individual sites.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge