Jingyuan Chou
Identification of Predictive Sub-Phenotypes of Acute Kidney Injury using Structured and Unstructured Electronic Health Record Data with Memory Networks
Apr 10, 2019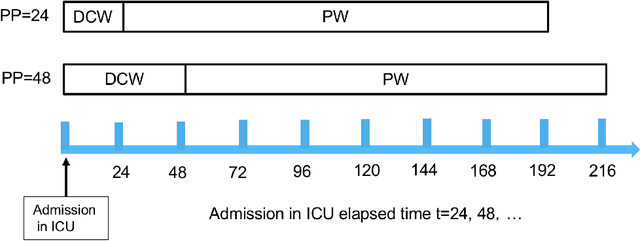
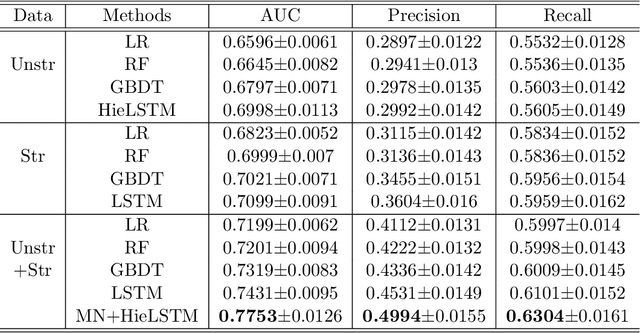
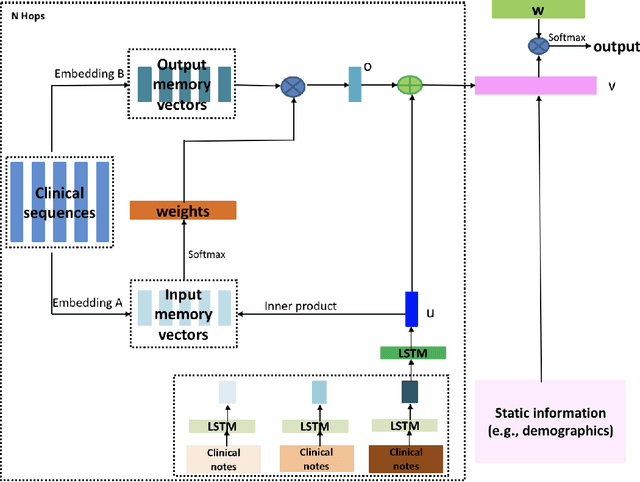
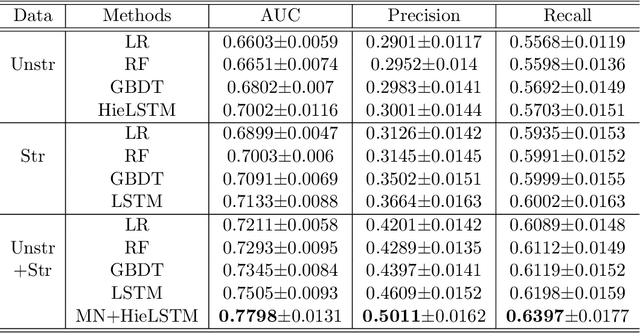
Abstract:Acute Kidney Injury (AKI) is a common clinical syndrome characterized by the rapid loss of kidney excretory function, which aggravates the clinical severity of other diseases in a large number of hospitalized patients. Accurate early prediction of AKI can enable in-time interventions and treatments. However, AKI is highly heterogeneous, thus identification of AKI sub-phenotypes can lead to an improved understanding of the disease pathophysiology and development of more targeted clinical interventions. This study used a memory network-based deep learning approach to discover predictive AKI sub-phenotypes using structured and unstructured electronic health record (EHR) data of patients before AKI diagnosis. We leveraged a real world critical care EHR corpus including 37,486 ICU stays. Our approach identified three distinct sub-phenotypes: sub-phenotype I is with an average age of 63.03$ \pm 17.25 $ years, and is characterized by mild loss of kidney excretory function (Serum Creatinne (SCr) $1.55\pm 0.34$ mg/dL, estimated Glomerular Filtration Rate Test (eGFR) $107.65\pm 54.98$ mL/min/1.73$m^2$). These patients are more likely to develop stage I AKI. Sub-phenotype II is with average age 66.81$ \pm 10.43 $ years, and was characterized by severe loss of kidney excretory function (SCr $1.96\pm 0.49$ mg/dL, eGFR $82.19\pm 55.92$ mL/min/1.73$m^2$). These patients are more likely to develop stage III AKI. Sub-phenotype III is with average age 65.07$ \pm 11.32 $ years, and was characterized moderate loss of kidney excretory function and thus more likely to develop stage II AKI (SCr $1.69\pm 0.32$ mg/dL, eGFR $93.97\pm 56.53$ mL/min/1.73$m^2$). Both SCr and eGFR are significantly different across the three sub-phenotypes with statistical testing plus postdoc analysis, and the conclusion still holds after age adjustment.
Integrative Analysis of Patient Health Records and Neuroimages via Memory-based Graph Convolutional Network
Oct 07, 2018


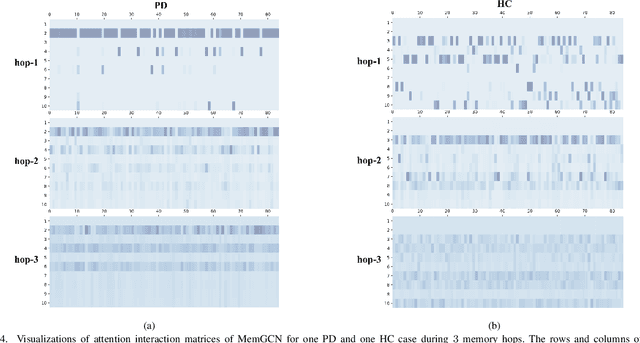
Abstract:With the arrival of the big data era, more and more data are becoming readily available in various real-world applications and those data are usually highly heterogeneous. Taking computational medicine as an example, we have both Electronic Health Records (EHR) and medical images for each patient. For complicated diseases such as Parkinson's and Alzheimer's, both EHR and neuroimaging information are very important for disease understanding because they contain complementary aspects of the disease. However, EHR and neuroimage are completely different. So far the existing research has been mainly focusing on one of them. In this paper, we proposed a framework, Memory-Based Graph Convolution Network (MemGCN), to perform integrative analysis with such multi-modal data. Specifically, GCN is used to extract useful information from the patients' neuroimages. The information contained in the patient EHRs before the acquisition of each brain image is captured by a memory network because of its sequential nature. The information contained in each brain image is combined with the information read out from the memory network to infer the disease state at the image acquisition timestamp. To further enhance the analytical power of MemGCN, we also designed a multi-hop strategy that allows multiple reading and updating on the memory can be performed at each iteration. We conduct experiments using the patient data from the Parkinson's Progression Markers Initiative (PPMI) with the task of classification of Parkinson's Disease (PD) cases versus controls. We demonstrate that superior classification performance can be achieved with our proposed framework, comparing with existing approaches involving a single type of data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge