Jyotishman Pathak
Extracting Social Support and Social Isolation Information from Clinical Psychiatry Notes: Comparing a Rule-based NLP System and a Large Language Model
Mar 25, 2024Abstract:Background: Social support (SS) and social isolation (SI) are social determinants of health (SDOH) associated with psychiatric outcomes. In electronic health records (EHRs), individual-level SS/SI is typically documented as narrative clinical notes rather than structured coded data. Natural language processing (NLP) algorithms can automate the otherwise labor-intensive process of data extraction. Data and Methods: Psychiatric encounter notes from Mount Sinai Health System (MSHS, n=300) and Weill Cornell Medicine (WCM, n=225) were annotated and established a gold standard corpus. A rule-based system (RBS) involving lexicons and a large language model (LLM) using FLAN-T5-XL were developed to identify mentions of SS and SI and their subcategories (e.g., social network, instrumental support, and loneliness). Results: For extracting SS/SI, the RBS obtained higher macro-averaged f-scores than the LLM at both MSHS (0.89 vs. 0.65) and WCM (0.85 vs. 0.82). For extracting subcategories, the RBS also outperformed the LLM at both MSHS (0.90 vs. 0.62) and WCM (0.82 vs. 0.81). Discussion and Conclusion: Unexpectedly, the RBS outperformed the LLMs across all metrics. Intensive review demonstrates that this finding is due to the divergent approach taken by the RBS and LLM. The RBS were designed and refined to follow the same specific rules as the gold standard annotations. Conversely, the LLM were more inclusive with categorization and conformed to common English-language understanding. Both approaches offer advantages and are made available open-source for future testing.
SODA: A Natural Language Processing Package to Extract Social Determinants of Health for Cancer Studies
Dec 06, 2022



Abstract:Objective: We aim to develop an open-source natural language processing (NLP) package, SODA (i.e., SOcial DeterminAnts), with pre-trained transformer models to extract social determinants of health (SDoH) for cancer patients, examine the generalizability of SODA to a new disease domain (i.e., opioid use), and evaluate the extraction rate of SDoH using cancer populations. Methods: We identified SDoH categories and attributes and developed an SDoH corpus using clinical notes from a general cancer cohort. We compared four transformer-based NLP models to extract SDoH, examined the generalizability of NLP models to a cohort of patients prescribed with opioids, and explored customization strategies to improve performance. We applied the best NLP model to extract 19 categories of SDoH from the breast (n=7,971), lung (n=11,804), and colorectal cancer (n=6,240) cohorts. Results and Conclusion: We developed a corpus of 629 cancer patients notes with annotations of 13,193 SDoH concepts/attributes from 19 categories of SDoH. The Bidirectional Encoder Representations from Transformers (BERT) model achieved the best strict/lenient F1 scores of 0.9216 and 0.9441 for SDoH concept extraction, 0.9617 and 0.9626 for linking attributes to SDoH concepts. Fine-tuning the NLP models using new annotations from opioid use patients improved the strict/lenient F1 scores from 0.8172/0.8502 to 0.8312/0.8679. The extraction rates among 19 categories of SDoH varied greatly, where 10 SDoH could be extracted from >70% of cancer patients, but 9 SDoH had a low extraction rate (<70% of cancer patients). The SODA package with pre-trained transformer models is publicly available at https://github.com/uf-hobiinformatics-lab/SDoH_SODA.
A Study of Social and Behavioral Determinants of Health in Lung Cancer Patients Using Transformers-based Natural Language Processing Models
Aug 10, 2021



Abstract:Social and behavioral determinants of health (SBDoH) have important roles in shaping people's health. In clinical research studies, especially comparative effectiveness studies, failure to adjust for SBDoH factors will potentially cause confounding issues and misclassification errors in either statistical analyses and machine learning-based models. However, there are limited studies to examine SBDoH factors in clinical outcomes due to the lack of structured SBDoH information in current electronic health record (EHR) systems, while much of the SBDoH information is documented in clinical narratives. Natural language processing (NLP) is thus the key technology to extract such information from unstructured clinical text. However, there is not a mature clinical NLP system focusing on SBDoH. In this study, we examined two state-of-the-art transformer-based NLP models, including BERT and RoBERTa, to extract SBDoH concepts from clinical narratives, applied the best performing model to extract SBDoH concepts on a lung cancer screening patient cohort, and examined the difference of SBDoH information between NLP extracted results and structured EHRs (SBDoH information captured in standard vocabularies such as the International Classification of Diseases codes). The experimental results show that the BERT-based NLP model achieved the best strict/lenient F1-score of 0.8791 and 0.8999, respectively. The comparison between NLP extracted SBDoH information and structured EHRs in the lung cancer patient cohort of 864 patients with 161,933 various types of clinical notes showed that much more detailed information about smoking, education, and employment were only captured in clinical narratives and that it is necessary to use both clinical narratives and structured EHRs to construct a more complete picture of patients' SBDoH factors.
Characterization of Time-variant and Time-invariant Assessment of Suicidality on Reddit using C-SSRS
Apr 09, 2021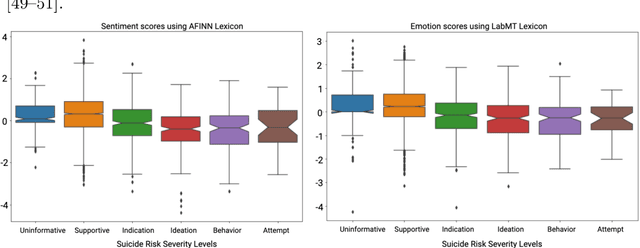
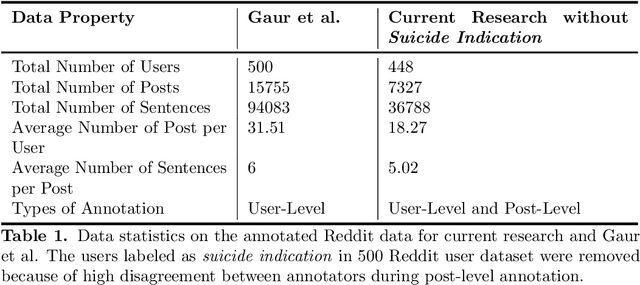
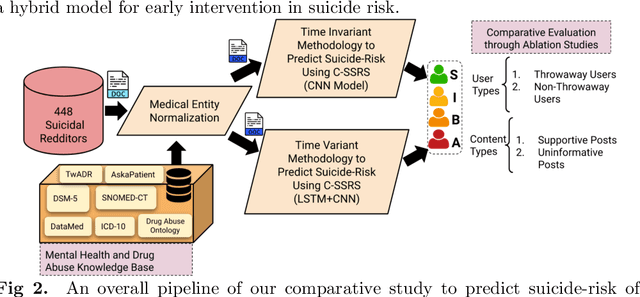
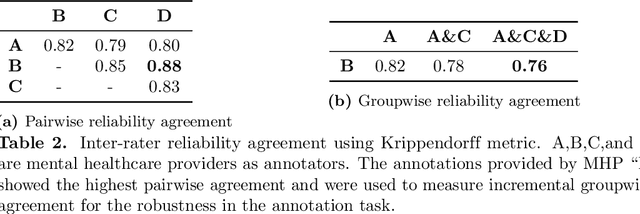
Abstract:Suicide is the 10th leading cause of death in the U.S (1999-2019). However, predicting when someone will attempt suicide has been nearly impossible. In the modern world, many individuals suffering from mental illness seek emotional support and advice on well-known and easily-accessible social media platforms such as Reddit. While prior artificial intelligence research has demonstrated the ability to extract valuable information from social media on suicidal thoughts and behaviors, these efforts have not considered both severity and temporality of risk. The insights made possible by access to such data have enormous clinical potential - most dramatically envisioned as a trigger to employ timely and targeted interventions (i.e., voluntary and involuntary psychiatric hospitalization) to save lives. In this work, we address this knowledge gap by developing deep learning algorithms to assess suicide risk in terms of severity and temporality from Reddit data based on the Columbia Suicide Severity Rating Scale (C-SSRS). In particular, we employ two deep learning approaches: time-variant and time-invariant modeling, for user-level suicide risk assessment, and evaluate their performance against a clinician-adjudicated gold standard Reddit corpus annotated based on the C-SSRS. Our results suggest that the time-variant approach outperforms the time-invariant method in the assessment of suicide-related ideations and supportive behaviors (AUC:0.78), while the time-invariant model performed better in predicting suicide-related behaviors and suicide attempt (AUC:0.64). The proposed approach can be integrated with clinical diagnostic interviews for improving suicide risk assessments.
A Machine Learning Approach to Detect Suicidal Ideation in US Veterans Based on Acoustic and Linguistic Features of Speech
Sep 27, 2020
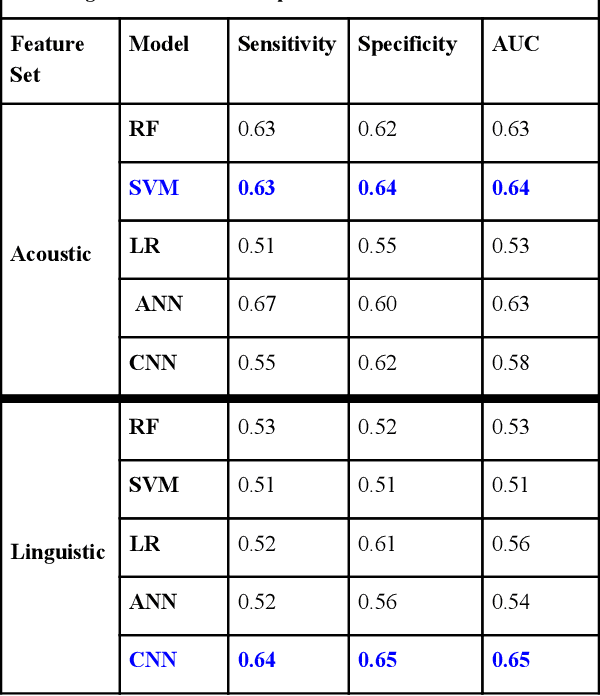
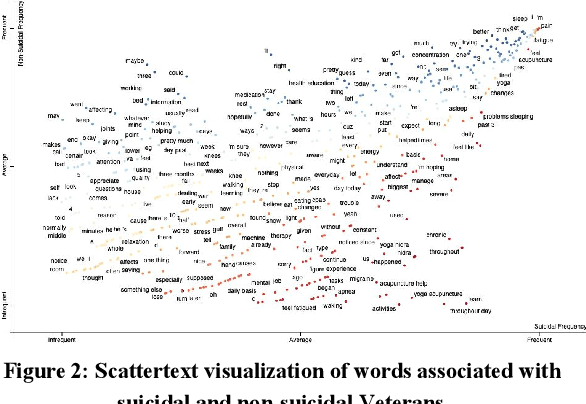
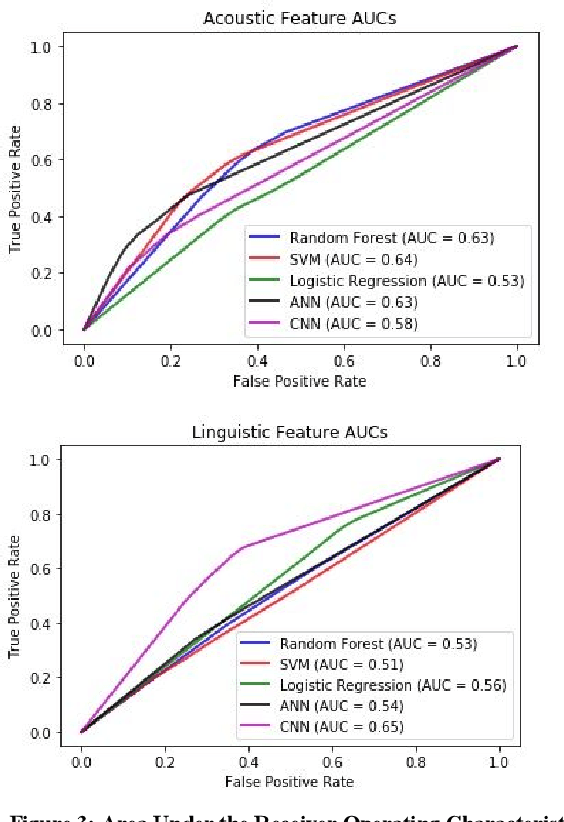
Abstract:Preventing Veteran suicide is a national priority. The US Department of Veterans Affairs (VA) collects, analyzes, and publishes data to inform suicide prevention strategies. Current approaches for detecting suicidal ideation mostly rely on patient self report which are inadequate and time consuming. In this research study, our goal was to automate suicidal ideation detection from acoustic and linguistic features of an individual's speech using machine learning (ML) algorithms. Using voice data collected from Veterans enrolled in a large interventional study on Gulf War Illness at the Washington DC VA Medical Center, we conducted an evaluation of the performance of different ML approaches in achieving our objective. By fitting both classical ML and deep learning models to the dataset, we identified the algorithms that were most effective for each feature set. Among classical machine learning algorithms, the Support Vector Machine (SVM) trained on acoustic features performed best in classifying suicidal Veterans. Among deep learning methods, the Convolutional Neural Network (CNN) trained on the linguistic features performed best. Our study shows that speech analysis in a machine learning pipeline is a promising approach for detecting suicidality among Veterans.
Identification of Predictive Sub-Phenotypes of Acute Kidney Injury using Structured and Unstructured Electronic Health Record Data with Memory Networks
Apr 10, 2019
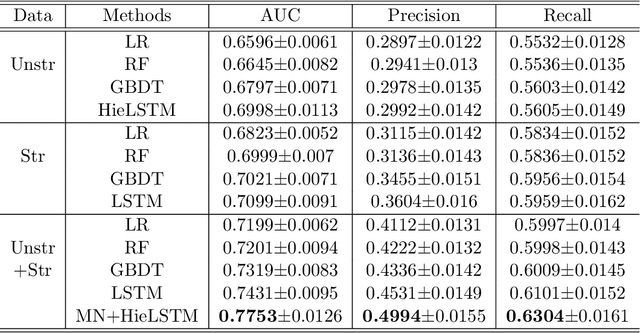
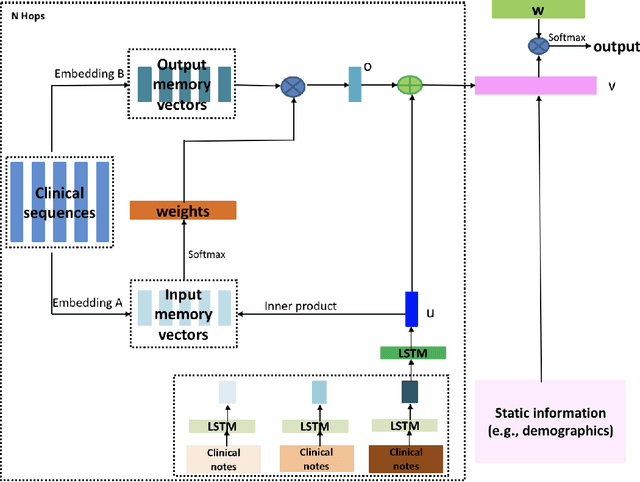
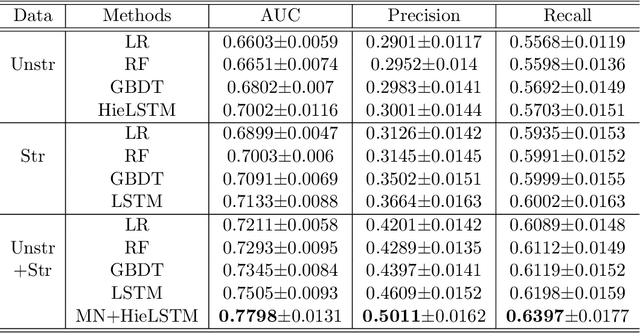
Abstract:Acute Kidney Injury (AKI) is a common clinical syndrome characterized by the rapid loss of kidney excretory function, which aggravates the clinical severity of other diseases in a large number of hospitalized patients. Accurate early prediction of AKI can enable in-time interventions and treatments. However, AKI is highly heterogeneous, thus identification of AKI sub-phenotypes can lead to an improved understanding of the disease pathophysiology and development of more targeted clinical interventions. This study used a memory network-based deep learning approach to discover predictive AKI sub-phenotypes using structured and unstructured electronic health record (EHR) data of patients before AKI diagnosis. We leveraged a real world critical care EHR corpus including 37,486 ICU stays. Our approach identified three distinct sub-phenotypes: sub-phenotype I is with an average age of 63.03$ \pm 17.25 $ years, and is characterized by mild loss of kidney excretory function (Serum Creatinne (SCr) $1.55\pm 0.34$ mg/dL, estimated Glomerular Filtration Rate Test (eGFR) $107.65\pm 54.98$ mL/min/1.73$m^2$). These patients are more likely to develop stage I AKI. Sub-phenotype II is with average age 66.81$ \pm 10.43 $ years, and was characterized by severe loss of kidney excretory function (SCr $1.96\pm 0.49$ mg/dL, eGFR $82.19\pm 55.92$ mL/min/1.73$m^2$). These patients are more likely to develop stage III AKI. Sub-phenotype III is with average age 65.07$ \pm 11.32 $ years, and was characterized moderate loss of kidney excretory function and thus more likely to develop stage II AKI (SCr $1.69\pm 0.32$ mg/dL, eGFR $93.97\pm 56.53$ mL/min/1.73$m^2$). Both SCr and eGFR are significantly different across the three sub-phenotypes with statistical testing plus postdoc analysis, and the conclusion still holds after age adjustment.
Evaluating the Portability of an NLP System for Processing Echocardiograms: A Retrospective, Multi-site Observational Study
Apr 02, 2019

Abstract:While natural language processing (NLP) of unstructured clinical narratives holds the potential for patient care and clinical research, portability of NLP approaches across multiple sites remains a major challenge. This study investigated the portability of an NLP system developed initially at the Department of Veterans Affairs (VA) to extract 27 key cardiac concepts from free-text or semi-structured echocardiograms from three academic medical centers: Weill Cornell Medicine, Mayo Clinic and Northwestern Medicine. While the NLP system showed high precision and recall measurements for four target concepts (aortic valve regurgitation, left atrium size at end systole, mitral valve regurgitation, tricuspid valve regurgitation) across all sites, we found moderate or poor results for the remaining concepts and the NLP system performance varied between individual sites.
Fusing Visual, Textual and Connectivity Clues for Studying Mental Health
Feb 19, 2019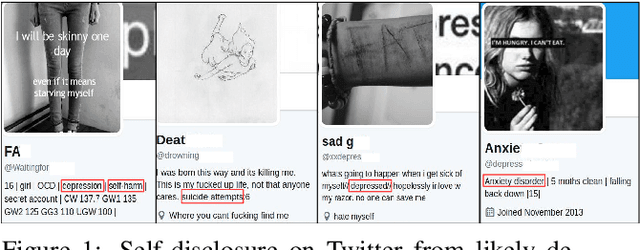
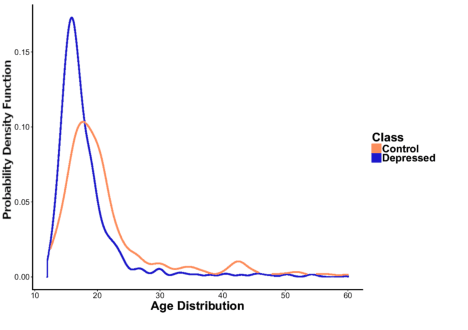
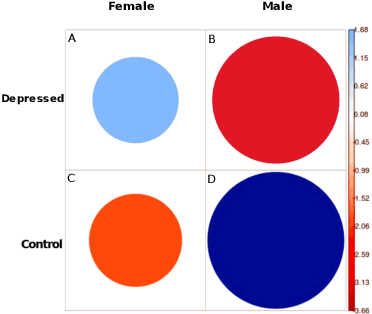
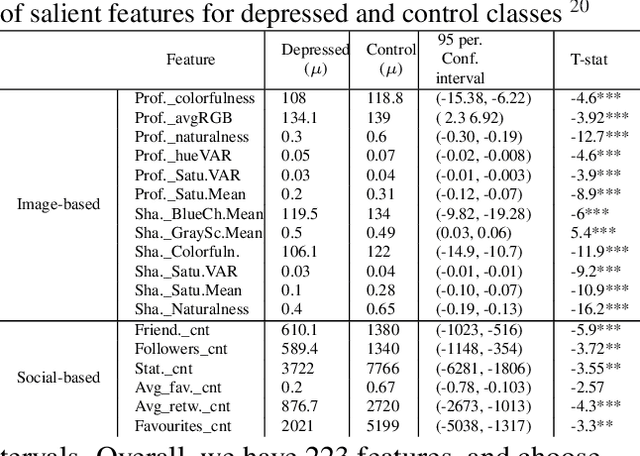
Abstract:With ubiquity of social media platforms, millions of people are sharing their online persona by expressing their thoughts, moods, emotions, feelings, and even their daily struggles with mental health issues voluntarily and publicly on social media. Unlike the most existing efforts which study depression by analyzing textual content, we examine and exploit multimodal big data to discern depressive behavior using a wide variety of features including individual-level demographics. By developing a multimodal framework and employing statistical techniques for fusing heterogeneous sets of features obtained by processing visual, textual and user interaction data, we significantly enhance the current state-of-the-art approaches for identifying depressed individuals on Twitter (improving the average F1-Score by 5 percent) as well as facilitate demographic inference from social media for broader applications. Besides providing insights into the relationship between demographics and mental health, our research assists in the design of a new breed of demographic-aware health interventions.
Developing a Portable Natural Language Processing Based Phenotyping System
Jul 17, 2018



Abstract:This paper presents a portable phenotyping system that is capable of integrating both rule-based and statistical machine learning based approaches. Our system utilizes UMLS to extract clinically relevant features from the unstructured text and then facilitates portability across different institutions and data systems by incorporating OHDSI's OMOP Common Data Model (CDM) to standardize necessary data elements. Our system can also store the key components of rule-based systems (e.g., regular expression matches) in the format of OMOP CDM, thus enabling the reuse, adaptation and extension of many existing rule-based clinical NLP systems. We experimented with our system on the corpus from i2b2's Obesity Challenge as a pilot study. Our system facilitates portable phenotyping of obesity and its 15 comorbidities based on the unstructured patient discharge summaries, while achieving a performance that often ranked among the top 10 of the challenge participants. This standardization enables a consistent application of numerous rule-based and machine learning based classification techniques downstream.
Semi-Supervised Approach to Monitoring Clinical Depressive Symptoms in Social Media
Oct 16, 2017



Abstract:With the rise of social media, millions of people are routinely expressing their moods, feelings, and daily struggles with mental health issues on social media platforms like Twitter. Unlike traditional observational cohort studies conducted through questionnaires and self-reported surveys, we explore the reliable detection of clinical depression from tweets obtained unobtrusively. Based on the analysis of tweets crawled from users with self-reported depressive symptoms in their Twitter profiles, we demonstrate the potential for detecting clinical depression symptoms which emulate the PHQ-9 questionnaire clinicians use today. Our study uses a semi-supervised statistical model to evaluate how the duration of these symptoms and their expression on Twitter (in terms of word usage patterns and topical preferences) align with the medical findings reported via the PHQ-9. Our proactive and automatic screening tool is able to identify clinical depressive symptoms with an accuracy of 68% and precision of 72%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge