Tanvi Banerjee
Wright State University
Debias-CLR: A Contrastive Learning Based Debiasing Method for Algorithmic Fairness in Healthcare Applications
Nov 20, 2024



Abstract:Artificial intelligence based predictive models trained on the clinical notes can be demographically biased. This could lead to adverse healthcare disparities in predicting outcomes like length of stay of the patients. Thus, it is necessary to mitigate the demographic biases within these models. We proposed an implicit in-processing debiasing method to combat disparate treatment which occurs when the machine learning model predict different outcomes for individuals based on the sensitive attributes like gender, ethnicity, race, and likewise. For this purpose, we used clinical notes of heart failure patients and used diagnostic codes, procedure reports and physiological vitals of the patients. We used Clinical BERT to obtain feature embeddings within the diagnostic codes and procedure reports, and LSTM autoencoders to obtain feature embeddings within the physiological vitals. Then, we trained two separate deep learning contrastive learning frameworks, one for gender and the other for ethnicity to obtain debiased representations within those demographic traits. We called this debiasing framework Debias-CLR. We leveraged clinical phenotypes of the patients identified in the diagnostic codes and procedure reports in the previous study to measure fairness statistically. We found that Debias-CLR was able to reduce the Single-Category Word Embedding Association Test (SC-WEAT) effect size score when debiasing for gender and ethnicity. We further found that to obtain fair representations in the embedding space using Debias-CLR, the accuracy of the predictive models on downstream tasks like predicting length of stay of the patients did not get reduced as compared to using the un-debiased counterparts for training the predictive models. Hence, we conclude that our proposed approach, Debias-CLR is fair and representative in mitigating demographic biases and can reduce health disparities.
Pain Forecasting using Self-supervised Learning and Patient Phenotyping: An attempt to prevent Opioid Addiction
Oct 09, 2023



Abstract:Sickle Cell Disease (SCD) is a chronic genetic disorder characterized by recurrent acute painful episodes. Opioids are often used to manage these painful episodes; the extent of their use in managing pain in this disorder is an issue of debate. The risk of addiction and side effects of these opioid treatments can often lead to more pain episodes in the future. Hence, it is crucial to forecast future patient pain trajectories to help patients manage their SCD to improve their quality of life without compromising their treatment. It is challenging to obtain many pain records to design forecasting models since it is mainly recorded by patients' self-report. Therefore, it is expensive and painful (due to the need for patient compliance) to solve pain forecasting problems in a purely supervised manner. In light of this challenge, we propose to solve the pain forecasting problem using self-supervised learning methods. Also, clustering such time-series data is crucial for patient phenotyping, anticipating patients' prognoses by identifying "similar" patients, and designing treatment guidelines tailored to homogeneous patient subgroups. Hence, we propose a self-supervised learning approach for clustering time-series data, where each cluster comprises patients who share similar future pain profiles. Experiments on five years of real-world datasets show that our models achieve superior performance over state-of-the-art benchmarks and identify meaningful clusters that can be translated into actionable information for clinical decision-making.
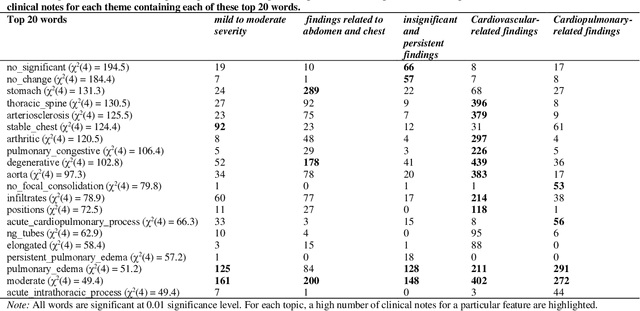
Mining Themes in Clinical Notes to Identify Phenotypes and to Predict Length of Stay in Patients admitted with Heart Failure
May 30, 2023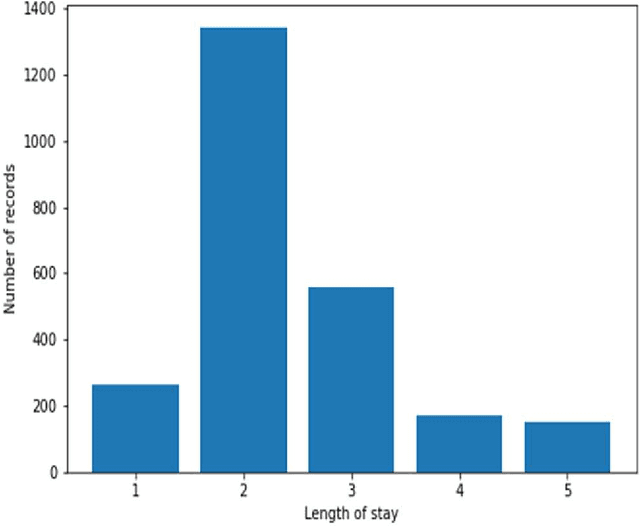
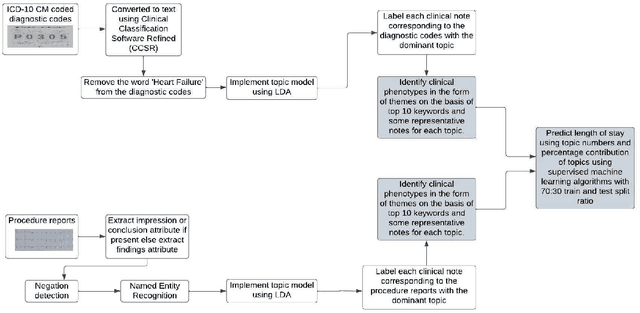
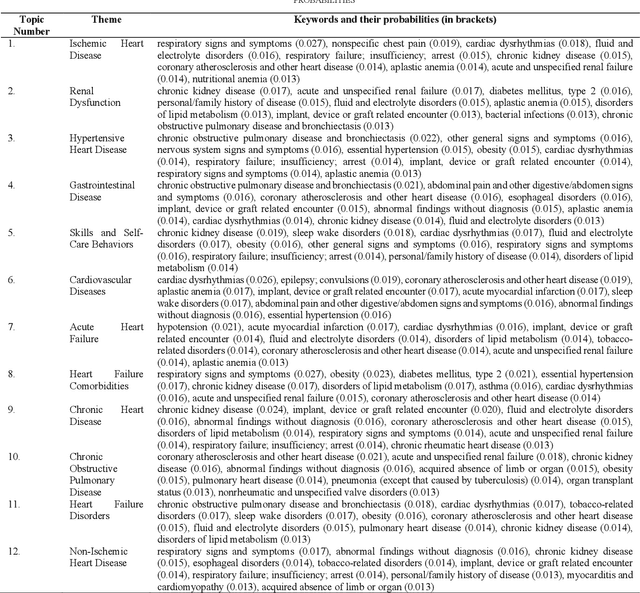
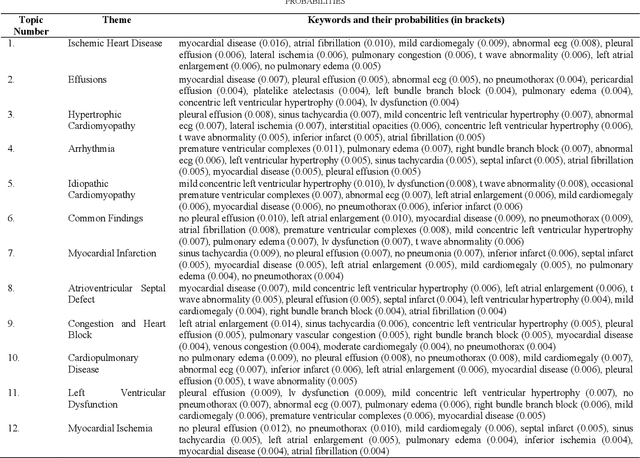
Abstract:Heart failure is a syndrome which occurs when the heart is not able to pump blood and oxygen to support other organs in the body. Identifying the underlying themes in the diagnostic codes and procedure reports of patients admitted for heart failure could reveal the clinical phenotypes associated with heart failure and to group patients based on their similar characteristics which could also help in predicting patient outcomes like length of stay. These clinical phenotypes usually have a probabilistic latent structure and hence, as there has been no previous work on identifying phenotypes in clinical notes of heart failure patients using a probabilistic framework and to predict length of stay of these patients using data-driven artificial intelligence-based methods, we apply natural language processing technique, topic modeling, to identify the themes present in diagnostic codes and in procedure reports of 1,200 patients admitted for heart failure at the University of Illinois Hospital and Health Sciences System (UI Health). Topic modeling identified twelve themes each in diagnostic codes and procedure reports which revealed information about different phenotypes related to various perspectives about heart failure, to study patients' profiles and to discover new relationships among medical concepts. Each theme had a set of keywords and each clinical note was labeled with two themes - one corresponding to its diagnostic code and the other corresponding to its procedure reports along with their percentage contribution. We used these themes and their percentage contribution to predict length of stay. We found that the themes discovered in diagnostic codes and procedure reports using topic modeling together were able to predict length of stay of the patients with an accuracy of 61.1% and an Area under the Receiver Operating Characteristic Curve (ROC AUC) value of 0.828.
Predicting Thermoelectric Power Factor of Bismuth Telluride During Laser Powder Bed Fusion Additive Manufacturing
Mar 28, 2023



Abstract:An additive manufacturing (AM) process, like laser powder bed fusion, allows for the fabrication of objects by spreading and melting powder in layers until a freeform part shape is created. In order to improve the properties of the material involved in the AM process, it is important to predict the material characterization property as a function of the processing conditions. In thermoelectric materials, the power factor is a measure of how efficiently the material can convert heat to electricity. While earlier works have predicted the material characterization properties of different thermoelectric materials using various techniques, implementation of machine learning models to predict the power factor of bismuth telluride (Bi2Te3) during the AM process has not been explored. This is important as Bi2Te3 is a standard material for low temperature applications. Thus, we used data about manufacturing processing parameters involved and in-situ sensor monitoring data collected during AM of Bi2Te3, to train different machine learning models in order to predict its thermoelectric power factor. We implemented supervised machine learning techniques using 80% training and 20% test data and further used the permutation feature importance method to identify important processing parameters and in-situ sensor features which were best at predicting power factor of the material. Ensemble-based methods like random forest, AdaBoost classifier, and bagging classifier performed the best in predicting power factor with the highest accuracy of 90% achieved by the bagging classifier model. Additionally, we found the top 15 processing parameters and in-situ sensor features to characterize the material manufacturing property like power factor. These features could further be optimized to maximize power factor of the thermoelectric material and improve the quality of the products built using this material.
Leveraging Natural Learning Processing to Uncover Themes in Clinical Notes of Patients Admitted for Heart Failure
Apr 14, 2022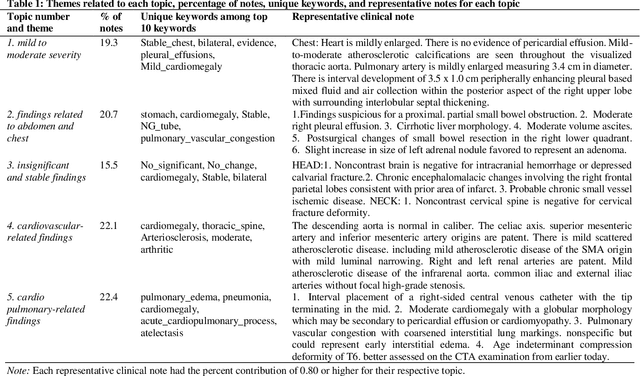
Abstract:Heart failure occurs when the heart is not able to pump blood and oxygen to support other organs in the body as it should. Treatments include medications and sometimes hospitalization. Patients with heart failure can have both cardiovascular as well as non-cardiovascular comorbidities. Clinical notes of patients with heart failure can be analyzed to gain insight into the topics discussed in these notes and the major comorbidities in these patients. In this regard, we apply machine learning techniques, such as topic modeling, to identify the major themes found in the clinical notes specific to the procedures performed on 1,200 patients admitted for heart failure at the University of Illinois Hospital and Health Sciences System (UI Health). Topic modeling revealed five hidden themes in these clinical notes, including one related to heart disease comorbidities.
Improving the Factual Accuracy of Abstractive Clinical Text Summarization using Multi-Objective Optimization
Apr 02, 2022
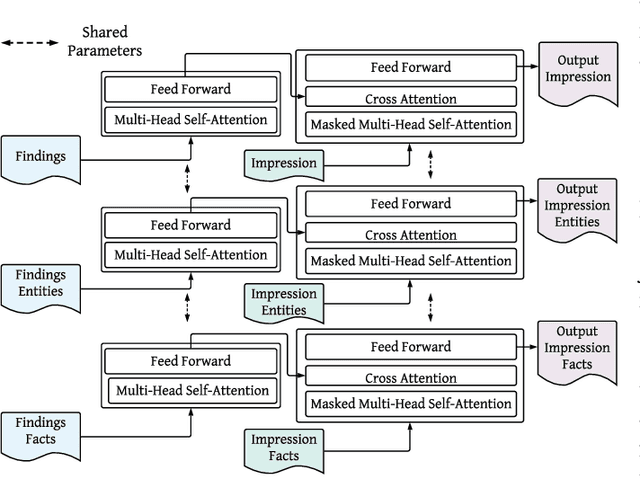
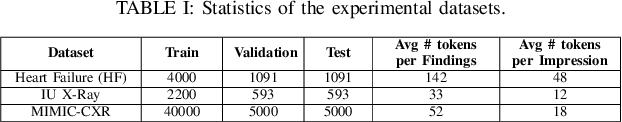
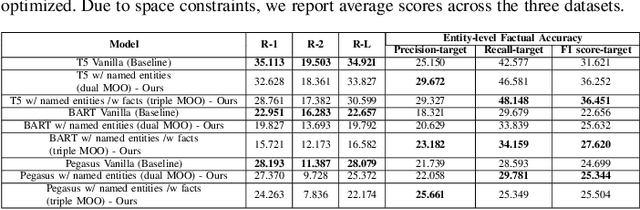
Abstract:While there has been recent progress in abstractive summarization as applied to different domains including news articles, scientific articles, and blog posts, the application of these techniques to clinical text summarization has been limited. This is primarily due to the lack of large-scale training data and the messy/unstructured nature of clinical notes as opposed to other domains where massive training data come in structured or semi-structured form. Further, one of the least explored and critical components of clinical text summarization is factual accuracy of clinical summaries. This is specifically crucial in the healthcare domain, cardiology in particular, where an accurate summary generation that preserves the facts in the source notes is critical to the well-being of a patient. In this study, we propose a framework for improving the factual accuracy of abstractive summarization of clinical text using knowledge-guided multi-objective optimization. We propose to jointly optimize three cost functions in our proposed architecture during training: generative loss, entity loss and knowledge loss and evaluate the proposed architecture on 1) clinical notes of patients with heart failure (HF), which we collect for this study; and 2) two benchmark datasets, Indiana University Chest X-ray collection (IU X-Ray), and MIMIC-CXR, that are publicly available. We experiment with three transformer encoder-decoder architectures and demonstrate that optimizing different loss functions leads to improved performance in terms of entity-level factual accuracy.
Entity-driven Fact-aware Abstractive Summarization of Biomedical Literature
Mar 30, 2022



Abstract:As part of the large number of scientific articles being published every year, the publication rate of biomedical literature has been increasing. Consequently, there has been considerable effort to harness and summarize the massive amount of biomedical research articles. While transformer-based encoder-decoder models in a vanilla source document-to-summary setting have been extensively studied for abstractive summarization in different domains, their major limitations continue to be entity hallucination (a phenomenon where generated summaries constitute entities not related to or present in source article(s)) and factual inconsistency. This problem is exacerbated in a biomedical setting where named entities and their semantics (which can be captured through a knowledge base) constitute the essence of an article. The use of named entities and facts mined from background knowledge bases pertaining to the named entities to guide abstractive summarization has not been studied in biomedical article summarization literature. In this paper, we propose an entity-driven fact-aware framework for training end-to-end transformer-based encoder-decoder models for abstractive summarization of biomedical articles. We call the proposed approach, whose building block is a transformer-based model, EFAS, Entity-driven Fact-aware Abstractive Summarization. We conduct experiments using five state-of-the-art transformer-based models (two of which are specifically designed for long document summarization) and demonstrate that injecting knowledge into the training/inference phase of these models enables the models to achieve significantly better performance than the standard source document-to-summary setting in terms of entity-level factual accuracy, N-gram novelty, and semantic equivalence while performing comparably on ROUGE metrics. The proposed approach is evaluated on ICD-11-Summ-1000, and PubMed-50k.
Clustering of Pain Dynamics in Sickle Cell Disease from Sparse, Uneven Samples
Aug 31, 2021



Abstract:Irregularly sampled time series data are common in a variety of fields. Many typical methods for drawing insight from data fail in this case. Here we attempt to generalize methods for clustering trajectories to irregularly and sparsely sampled data. We first construct synthetic data sets, then propose and assess four methods of data alignment to allow for application of spectral clustering. We also repeat the same process for real data drawn from medical records of patients with sickle cell disease -- patients whose subjective experiences of pain were tracked for several months via a mobile app. We find that different methods for aligning irregularly sampled sparse data sets can lead to different optimal numbers of clusters, even for synthetic data with known properties. For the case of sickle cell disease, we find that three clusters is a reasonable choice, and these appear to correspond to (1) a low pain group with occasionally acute pain, (2) a group which experiences moderate mean pain that fluctuates often from low to high, and (3) a group that experiences persistent high levels of pain. Our results may help physicians and patients better understand and manage patients' pain levels over time, and we expect that the methods we develop will apply to a wide range of other data sources in medicine and beyond.
Pain Intensity Assessment in Sickle Cell Disease patients using Vital Signs during Hospital Visits
Nov 24, 2020


Abstract:Pain in sickle cell disease (SCD) is often associated with increased morbidity, mortality, and high healthcare costs. The standard method for predicting the absence, presence, and intensity of pain has long been self-report. However, medical providers struggle to manage patients based on subjective pain reports correctly and pain medications often lead to further difficulties in patient communication as they may cause sedation and sleepiness. Recent studies have shown that objective physiological measures can predict subjective self-reported pain scores for inpatient visits using machine learning (ML) techniques. In this study, we evaluate the generalizability of ML techniques to data collected from 50 patients over an extended period across three types of hospital visits (i.e., inpatient, outpatient and outpatient evaluation). We compare five classification algorithms for various pain intensity levels at both intra-individual (within each patient) and inter-individual (between patients) level. While all the tested classifiers perform much better than chance, a Decision Tree (DT) model performs best at predicting pain on an 11-point severity scale (from 0-10) with an accuracy of 0.728 at an inter-individual level and 0.653 at an intra-individual level. The accuracy of DT significantly improves to 0.941 on a 2-point rating scale (i.e., no/mild pain: 0-5, severe pain: 6-10) at an intra-individual level. Our experimental results demonstrate that ML techniques can provide an objective and quantitative evaluation of pain intensity levels for all three types of hospital visits.
COVID-19 and Mental Health/Substance Use Disorders on Reddit: A Longitudinal Study
Nov 20, 2020



Abstract:COVID-19 pandemic has adversely and disproportionately impacted people suffering from mental health issues and substance use problems. This has been exacerbated by social isolation during the pandemic and the social stigma associated with mental health and substance use disorders, making people reluctant to share their struggles and seek help. Due to the anonymity and privacy they provide, social media emerged as a convenient medium for people to share their experiences about their day to day struggles. Reddit is a well-recognized social media platform that provides focused and structured forums called subreddits, that users subscribe to and discuss their experiences with others. Temporal assessment of the topical correlation between social media postings about mental health/substance use and postings about Coronavirus is crucial to better understand public sentiment on the pandemic and its evolving impact, especially related to vulnerable populations. In this study, we conduct a longitudinal topical analysis of postings between subreddits r/depression, r/Anxiety, r/SuicideWatch, and r/Coronavirus, and postings between subreddits r/opiates, r/OpiatesRecovery, r/addiction, and r/Coronavirus from January 2020 - October 2020. Our results show a high topical correlation between postings in r/depression and r/Coronavirus in September 2020. Further, the topical correlation between postings on substance use disorders and Coronavirus fluctuates, showing the highest correlation in August 2020. By monitoring these trends from platforms such as Reddit, epidemiologists, and mental health professionals can gain insights into the challenges faced by communities for targeted interventions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge