Luke Rasmussen
Characterizing Design Patterns of EHR-Driven Phenotype Extraction Algorithms
Nov 15, 2018
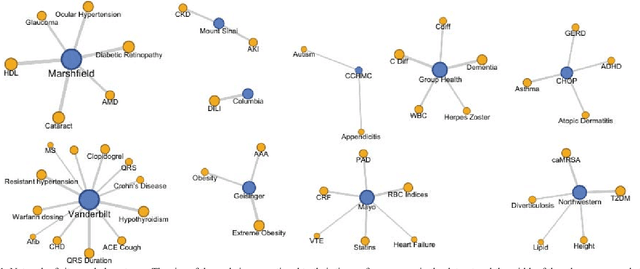
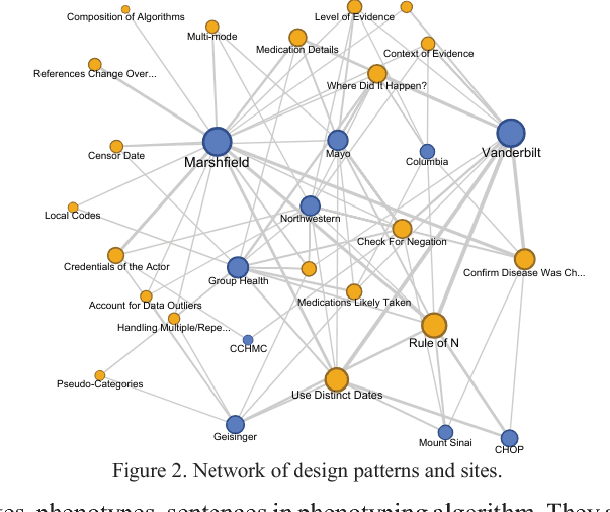
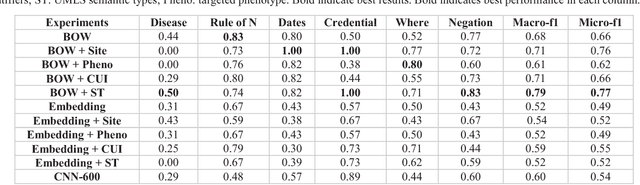
Abstract:The automatic development of phenotype algorithms from Electronic Health Record data with machine learning (ML) techniques is of great interest given the current practice is very time-consuming and resource intensive. The extraction of design patterns from phenotype algorithms is essential to understand their rationale and standard, with great potential to automate the development process. In this pilot study, we perform network visualization on the design patterns and their associations with phenotypes and sites. We classify design patterns using the fragments from previously annotated phenotype algorithms as the ground truth. The classification performance is used as a proxy for coherence at the attribution level. The bag-of-words representation with knowledge-based features generated a good performance in the classification task (0.79 macro-f1 scores). Good classification accuracy with simple features demonstrated the attribution coherence and the feasibility of automatic identification of design patterns. Our results point to both the feasibility and challenges of automatic identification of phenotyping design patterns, which would power the automatic development of phenotype algorithms.
Developing a Portable Natural Language Processing Based Phenotyping System
Jul 17, 2018



Abstract:This paper presents a portable phenotyping system that is capable of integrating both rule-based and statistical machine learning based approaches. Our system utilizes UMLS to extract clinically relevant features from the unstructured text and then facilitates portability across different institutions and data systems by incorporating OHDSI's OMOP Common Data Model (CDM) to standardize necessary data elements. Our system can also store the key components of rule-based systems (e.g., regular expression matches) in the format of OMOP CDM, thus enabling the reuse, adaptation and extension of many existing rule-based clinical NLP systems. We experimented with our system on the corpus from i2b2's Obesity Challenge as a pilot study. Our system facilitates portable phenotyping of obesity and its 15 comorbidities based on the unstructured patient discharge summaries, while achieving a performance that often ranked among the top 10 of the challenge participants. This standardization enables a consistent application of numerous rule-based and machine learning based classification techniques downstream.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge