Yizhen Zhong
Improving Precancerous Case Characterization via Transformer-based Ensemble Learning
Dec 10, 2022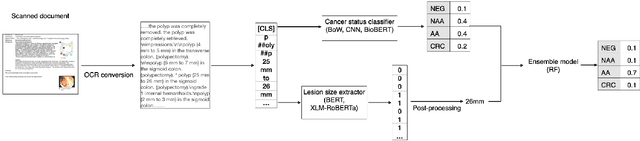

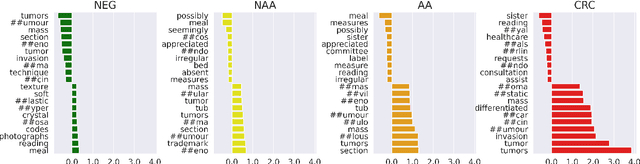
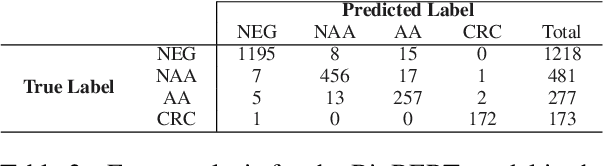
Abstract:The application of natural language processing (NLP) to cancer pathology reports has been focused on detecting cancer cases, largely ignoring precancerous cases. Improving the characterization of precancerous adenomas assists in developing diagnostic tests for early cancer detection and prevention, especially for colorectal cancer (CRC). Here we developed transformer-based deep neural network NLP models to perform the CRC phenotyping, with the goal of extracting precancerous lesion attributes and distinguishing cancer and precancerous cases. We achieved 0.914 macro-F1 scores for classifying patients into negative, non-advanced adenoma, advanced adenoma and CRC. We further improved the performance to 0.923 using an ensemble of classifiers for cancer status classification and lesion size named entity recognition (NER). Our results demonstrated the potential of using NLP to leverage real-world health record data to facilitate the development of diagnostic tests for early cancer prevention.
Evaluating the Portability of an NLP System for Processing Echocardiograms: A Retrospective, Multi-site Observational Study
Apr 02, 2019

Abstract:While natural language processing (NLP) of unstructured clinical narratives holds the potential for patient care and clinical research, portability of NLP approaches across multiple sites remains a major challenge. This study investigated the portability of an NLP system developed initially at the Department of Veterans Affairs (VA) to extract 27 key cardiac concepts from free-text or semi-structured echocardiograms from three academic medical centers: Weill Cornell Medicine, Mayo Clinic and Northwestern Medicine. While the NLP system showed high precision and recall measurements for four target concepts (aortic valve regurgitation, left atrium size at end systole, mitral valve regurgitation, tricuspid valve regurgitation) across all sites, we found moderate or poor results for the remaining concepts and the NLP system performance varied between individual sites.
Characterizing Design Patterns of EHR-Driven Phenotype Extraction Algorithms
Nov 15, 2018
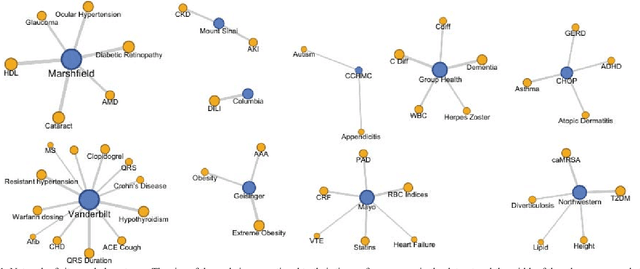
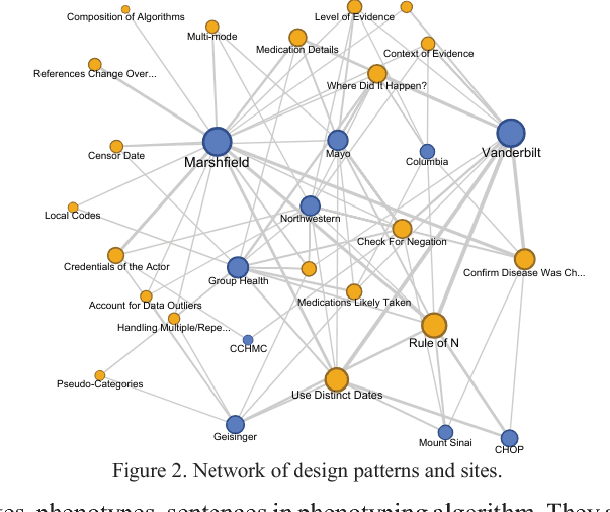
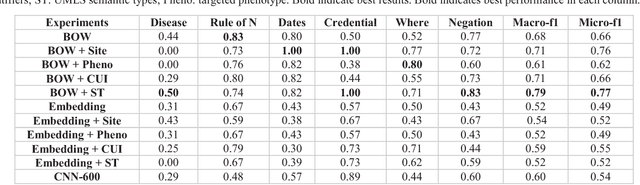
Abstract:The automatic development of phenotype algorithms from Electronic Health Record data with machine learning (ML) techniques is of great interest given the current practice is very time-consuming and resource intensive. The extraction of design patterns from phenotype algorithms is essential to understand their rationale and standard, with great potential to automate the development process. In this pilot study, we perform network visualization on the design patterns and their associations with phenotypes and sites. We classify design patterns using the fragments from previously annotated phenotype algorithms as the ground truth. The classification performance is used as a proxy for coherence at the attribution level. The bag-of-words representation with knowledge-based features generated a good performance in the classification task (0.79 macro-f1 scores). Good classification accuracy with simple features demonstrated the attribution coherence and the feasibility of automatic identification of design patterns. Our results point to both the feasibility and challenges of automatic identification of phenotyping design patterns, which would power the automatic development of phenotype algorithms.
Developing a Portable Natural Language Processing Based Phenotyping System
Jul 17, 2018



Abstract:This paper presents a portable phenotyping system that is capable of integrating both rule-based and statistical machine learning based approaches. Our system utilizes UMLS to extract clinically relevant features from the unstructured text and then facilitates portability across different institutions and data systems by incorporating OHDSI's OMOP Common Data Model (CDM) to standardize necessary data elements. Our system can also store the key components of rule-based systems (e.g., regular expression matches) in the format of OMOP CDM, thus enabling the reuse, adaptation and extension of many existing rule-based clinical NLP systems. We experimented with our system on the corpus from i2b2's Obesity Challenge as a pilot study. Our system facilitates portable phenotyping of obesity and its 15 comorbidities based on the unstructured patient discharge summaries, while achieving a performance that often ranked among the top 10 of the challenge participants. This standardization enables a consistent application of numerous rule-based and machine learning based classification techniques downstream.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge