Goonmeet Bajaj
Grounding from an AI and Cognitive Science Lens
Feb 19, 2024Abstract:Grounding is a challenging problem, requiring a formal definition and different levels of abstraction. This article explores grounding from both cognitive science and machine learning perspectives. It identifies the subtleties of grounding, its significance for collaborative agents, and similarities and differences in grounding approaches in both communities. The article examines the potential of neuro-symbolic approaches tailored for grounding tasks, showcasing how they can more comprehensively address grounding. Finally, we discuss areas for further exploration and development in grounding.
UBERT: A Novel Language Model for Synonymy Prediction at Scale in the UMLS Metathesaurus
Apr 27, 2022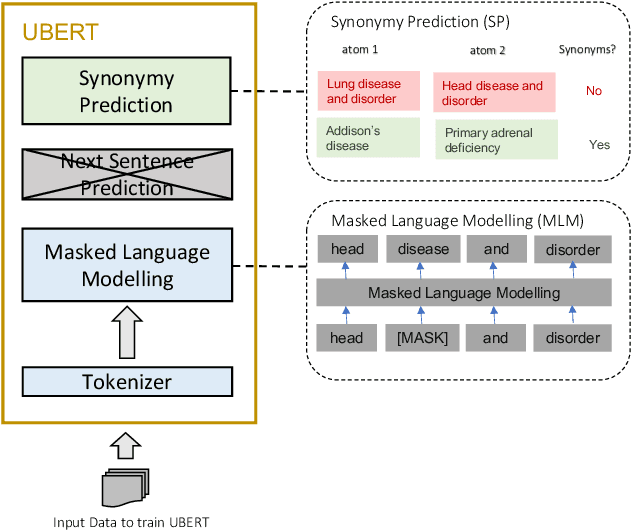

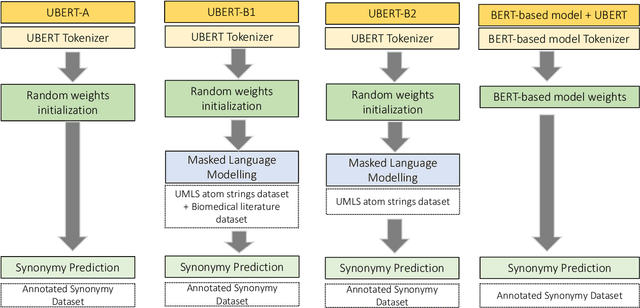

Abstract:The UMLS Metathesaurus integrates more than 200 biomedical source vocabularies. During the Metathesaurus construction process, synonymous terms are clustered into concepts by human editors, assisted by lexical similarity algorithms. This process is error-prone and time-consuming. Recently, a deep learning model (LexLM) has been developed for the UMLS Vocabulary Alignment (UVA) task. This work introduces UBERT, a BERT-based language model, pretrained on UMLS terms via a supervised Synonymy Prediction (SP) task replacing the original Next Sentence Prediction (NSP) task. The effectiveness of UBERT for UMLS Metathesaurus construction process is evaluated using the UMLS Vocabulary Alignment (UVA) task. We show that UBERT outperforms the LexLM, as well as biomedical BERT-based models. Key to the performance of UBERT are the synonymy prediction task specifically developed for UBERT, the tight alignment of training data to the UVA task, and the similarity of the models used for pretrained UBERT.
Evaluating Biomedical BERT Models for Vocabulary Alignment at Scale in the UMLS Metathesaurus
Sep 14, 2021
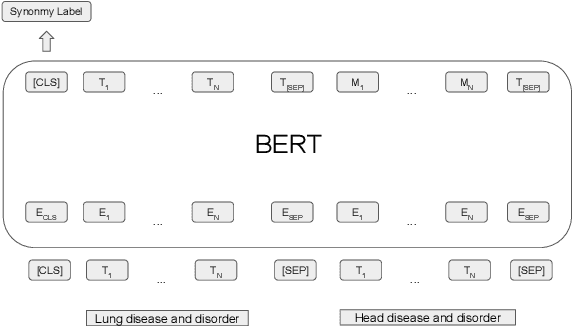

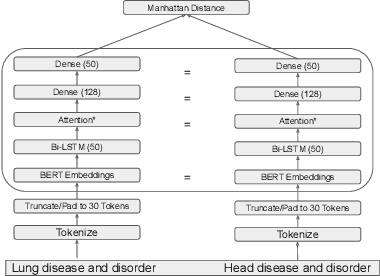
Abstract:The current UMLS (Unified Medical Language System) Metathesaurus construction process for integrating over 200 biomedical source vocabularies is expensive and error-prone as it relies on the lexical algorithms and human editors for deciding if the two biomedical terms are synonymous. Recent advances in Natural Language Processing such as Transformer models like BERT and its biomedical variants with contextualized word embeddings have achieved state-of-the-art (SOTA) performance on downstream tasks. We aim to validate if these approaches using the BERT models can actually outperform the existing approaches for predicting synonymy in the UMLS Metathesaurus. In the existing Siamese Networks with LSTM and BioWordVec embeddings, we replace the BioWordVec embeddings with the biomedical BERT embeddings extracted from each BERT model using different ways of extraction. In the Transformer architecture, we evaluate the use of the different biomedical BERT models that have been pre-trained using different datasets and tasks. Given the SOTA performance of these BERT models for other downstream tasks, our experiments yield surprisingly interesting results: (1) in both model architectures, the approaches employing these biomedical BERT-based models do not outperform the existing approaches using Siamese Network with BioWordVec embeddings for the UMLS synonymy prediction task, (2) the original BioBERT large model that has not been pre-trained with the UMLS outperforms the SapBERT models that have been pre-trained with the UMLS, and (3) using the Siamese Networks yields better performance for synonymy prediction when compared to using the biomedical BERT models.
Understanding Knowledge Gaps in Visual Question Answering: Implications for Gap Identification and Testing
Apr 08, 2020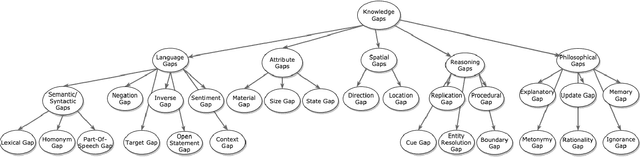
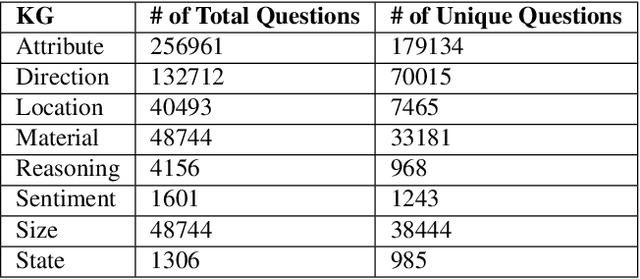

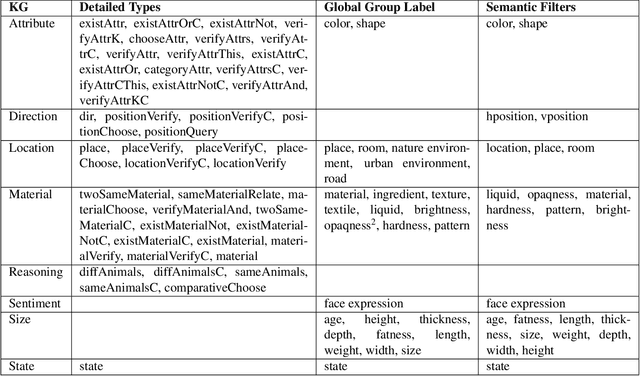
Abstract:Visual Question Answering (VQA) systems are tasked with answering natural language questions corresponding to a presented image. Current VQA datasets typically contain questions related to the spatial information of objects, object attributes, or general scene questions. Recently, researchers have recognized the need for improving the balance of such datasets to reduce the system's dependency on memorized linguistic features and statistical biases and to allow for improved visual understanding. However, it is unclear as to whether there are any latent patterns that can be used to quantify and explain these failures. To better quantify our understanding of the performance of VQA models, we use a taxonomy of Knowledge Gaps (KGs) to identify/tag questions with one or more types of KGs. Each KG describes the reasoning abilities needed to arrive at a resolution, and failure to resolve gaps indicate an absence of the required reasoning ability. After identifying KGs for each question, we examine the skew in the distribution of the number of questions for each KG. In order to reduce the skew in the distribution of questions across KGs, we introduce a targeted question generation model. This model allows us to generate new types of questions for an image.
Network Representation Learning: Consolidation and Renewed Bearing
May 02, 2019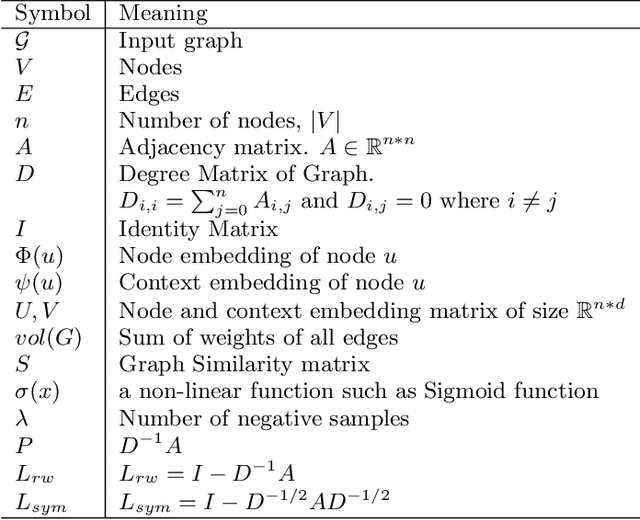
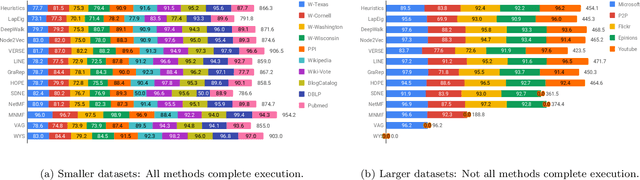

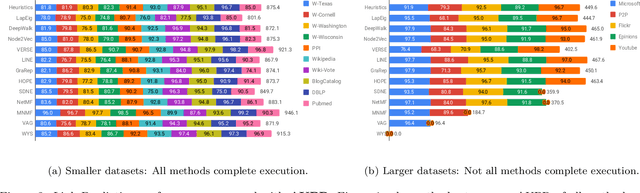
Abstract:Graphs are a natural abstraction for many problems where nodes represent entities and edges represent a relationship across entities. An important area of research that has emerged over the last decade is the use of graphs as a vehicle for non-linear dimensionality reduction in a manner akin to previous efforts based on manifold learning with uses for downstream database processing, machine learning and visualization. In this systematic yet comprehensive experimental survey, we benchmark several popular network representation learning methods operating on two key tasks: link prediction and node classification. We examine the performance of 12 unsupervised embedding methods on 15 datasets. To the best of our knowledge, the scale of our study -- both in terms of the number of methods and number of datasets -- is the largest to date. Our results reveal several key insights about work-to-date in this space. First, we find that certain baseline methods (task-specific heuristics, as well as classic manifold methods) that have often been dismissed or are not considered by previous efforts can compete on certain types of datasets if they are tuned appropriately. Second, we find that recent methods based on matrix factorization offer a small but relatively consistent advantage over alternative methods (e.g., random-walk based methods) from a qualitative standpoint. Specifically, we find that MNMF, a community preserving embedding method, is the most competitive method for the link prediction task. While NetMF is the most competitive baseline for node classification. Third, no single method completely outperforms other embedding methods on both node classification and link prediction tasks. We also present several drill-down analysis that reveals settings under which certain algorithms perform well (e.g., the role of neighborhood context on performance) -- guiding the end-user.
Fusing Visual, Textual and Connectivity Clues for Studying Mental Health
Feb 19, 2019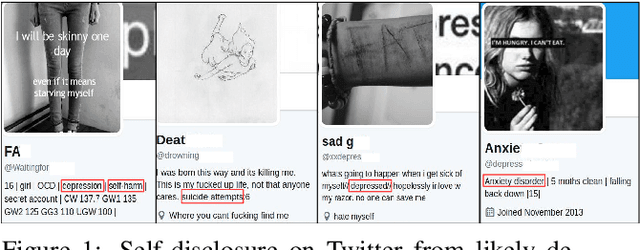
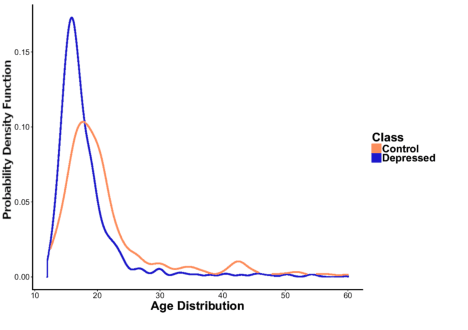
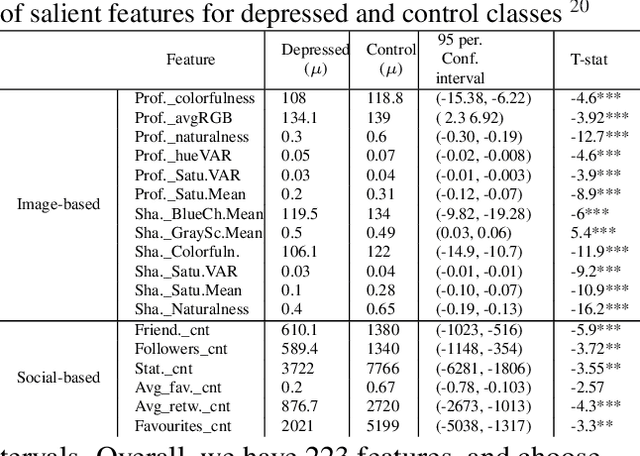
Abstract:With ubiquity of social media platforms, millions of people are sharing their online persona by expressing their thoughts, moods, emotions, feelings, and even their daily struggles with mental health issues voluntarily and publicly on social media. Unlike the most existing efforts which study depression by analyzing textual content, we examine and exploit multimodal big data to discern depressive behavior using a wide variety of features including individual-level demographics. By developing a multimodal framework and employing statistical techniques for fusing heterogeneous sets of features obtained by processing visual, textual and user interaction data, we significantly enhance the current state-of-the-art approaches for identifying depressed individuals on Twitter (improving the average F1-Score by 5 percent) as well as facilitate demographic inference from social media for broader applications. Besides providing insights into the relationship between demographics and mental health, our research assists in the design of a new breed of demographic-aware health interventions.
Semi-Supervised Approach to Monitoring Clinical Depressive Symptoms in Social Media
Oct 16, 2017



Abstract:With the rise of social media, millions of people are routinely expressing their moods, feelings, and daily struggles with mental health issues on social media platforms like Twitter. Unlike traditional observational cohort studies conducted through questionnaires and self-reported surveys, we explore the reliable detection of clinical depression from tweets obtained unobtrusively. Based on the analysis of tweets crawled from users with self-reported depressive symptoms in their Twitter profiles, we demonstrate the potential for detecting clinical depression symptoms which emulate the PHQ-9 questionnaire clinicians use today. Our study uses a semi-supervised statistical model to evaluate how the duration of these symptoms and their expression on Twitter (in terms of word usage patterns and topical preferences) align with the medical findings reported via the PHQ-9. Our proactive and automatic screening tool is able to identify clinical depressive symptoms with an accuracy of 68% and precision of 72%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge