Thilini Wijesiriwardene
Exploring the Abilities of Large Language Models to Solve Proportional Analogies via Knowledge-Enhanced Prompting
Dec 01, 2024



Abstract:Making analogies is fundamental to cognition. Proportional analogies, which consist of four terms, are often used to assess linguistic and cognitive abilities. For instance, completing analogies like "Oxygen is to Gas as <blank> is to <blank>" requires identifying the semantic relationship (e.g., "type of") between the first pair of terms ("Oxygen" and "Gas") and finding a second pair that shares the same relationship (e.g., "Aluminum" and "Metal"). In this work, we introduce a 15K Multiple-Choice Question Answering (MCQA) dataset for proportional analogy completion and evaluate the performance of contemporary Large Language Models (LLMs) in various knowledge-enhanced prompt settings. Specifically, we augment prompts with three types of knowledge: exemplar, structured, and targeted. Our results show that despite extensive training data, solving proportional analogies remains challenging for current LLMs, with the best model achieving an accuracy of 55%. Notably, we find that providing targeted knowledge can better assist models in completing proportional analogies compared to providing exemplars or collections of structured knowledge.
Exploring the Relationship between Analogy Identification and Sentence Structure Encoding in Large Language Models
Oct 13, 2023Abstract:Identifying analogies plays a pivotal role in human cognition and language proficiency. In the last decade, there has been extensive research on word analogies in the form of ``A is to B as C is to D.'' However, there is a growing interest in analogies that involve longer text, such as sentences and collections of sentences, which convey analogous meanings. While the current NLP research community evaluates the ability of Large Language Models (LLMs) to identify such analogies, the underlying reasons behind these abilities warrant deeper investigation. Furthermore, the capability of LLMs to encode both syntactic and semantic structures of language within their embeddings has garnered significant attention with the surge in their utilization. In this work, we examine the relationship between the abilities of multiple LLMs to identify sentence analogies, and their capacity to encode syntactic and semantic structures. Through our analysis, we find that analogy identification ability of LLMs is positively correlated with their ability to encode syntactic and semantic structures of sentences. Specifically, we find that the LLMs which capture syntactic structures better, also have higher abilities in identifying sentence analogies.
Why Do We Need Neuro-symbolic AI to Model Pragmatic Analogies?
Aug 02, 2023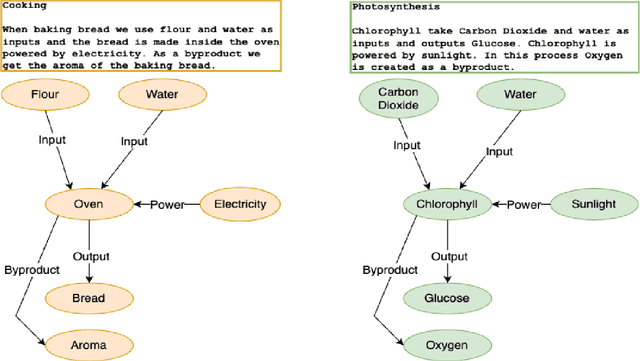
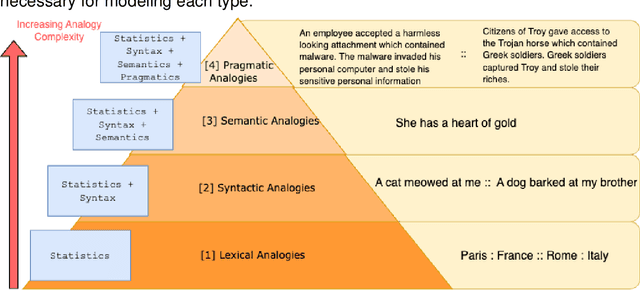
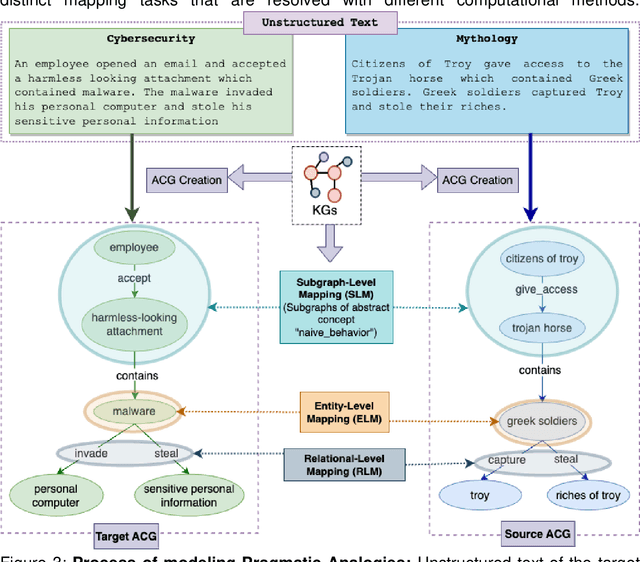
Abstract:A hallmark of intelligence is the ability to use a familiar domain to make inferences about a less familiar domain, known as analogical reasoning. In this article, we delve into the performance of Large Language Models (LLMs) in dealing with progressively complex analogies expressed in unstructured text. We discuss analogies at four distinct levels of complexity: lexical analogies, syntactic analogies, semantic analogies, and pragmatic analogies. As the analogies become more complex, they require increasingly extensive, diverse knowledge beyond the textual content, unlikely to be found in the lexical co-occurrence statistics that power LLMs. To address this, we discuss the necessity of employing Neuro-symbolic AI techniques that combine statistical and symbolic AI, informing the representation of unstructured text to highlight and augment relevant content, provide abstraction and guide the mapping process. Our knowledge-informed approach maintains the efficiency of LLMs while preserving the ability to explain analogies for pedagogical applications.
ANALOGICAL -- A New Benchmark for Analogy of Long Text for Large Language Models
May 14, 2023



Abstract:Over the past decade, analogies, in the form of word-level analogies, have played a significant role as an intrinsic measure of evaluating the quality of word embedding methods such as word2vec. Modern large language models (LLMs), however, are primarily evaluated on extrinsic measures based on benchmarks such as GLUE and SuperGLUE, and there are only a few investigations on whether LLMs can draw analogies between long texts. In this paper, we present ANALOGICAL, a new benchmark to intrinsically evaluate LLMs across a taxonomy of analogies of long text with six levels of complexity -- (i) word, (ii) word vs. sentence, (iii) syntactic, (iv) negation, (v) entailment, and (vi) metaphor. Using thirteen datasets and three different distance measures, we evaluate the abilities of eight LLMs in identifying analogical pairs in the semantic vector space. Our evaluation finds that it is increasingly challenging for LLMs to identify analogies when going up the analogy taxonomy.
UBERT: A Novel Language Model for Synonymy Prediction at Scale in the UMLS Metathesaurus
Apr 27, 2022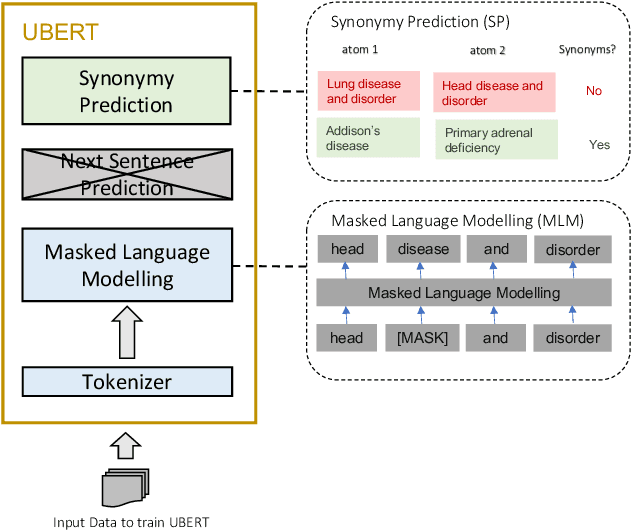

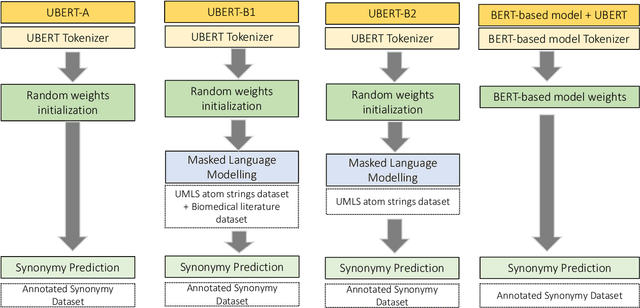

Abstract:The UMLS Metathesaurus integrates more than 200 biomedical source vocabularies. During the Metathesaurus construction process, synonymous terms are clustered into concepts by human editors, assisted by lexical similarity algorithms. This process is error-prone and time-consuming. Recently, a deep learning model (LexLM) has been developed for the UMLS Vocabulary Alignment (UVA) task. This work introduces UBERT, a BERT-based language model, pretrained on UMLS terms via a supervised Synonymy Prediction (SP) task replacing the original Next Sentence Prediction (NSP) task. The effectiveness of UBERT for UMLS Metathesaurus construction process is evaluated using the UMLS Vocabulary Alignment (UVA) task. We show that UBERT outperforms the LexLM, as well as biomedical BERT-based models. Key to the performance of UBERT are the synonymy prediction task specifically developed for UBERT, the tight alignment of training data to the UVA task, and the similarity of the models used for pretrained UBERT.
Evaluating Biomedical BERT Models for Vocabulary Alignment at Scale in the UMLS Metathesaurus
Sep 14, 2021
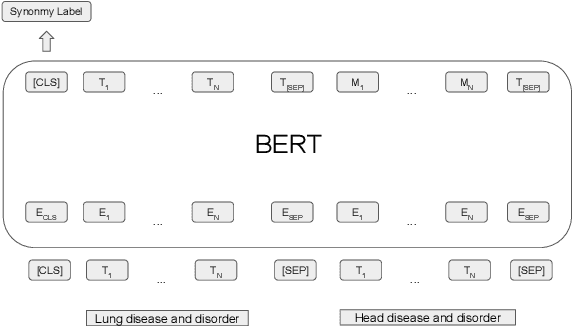

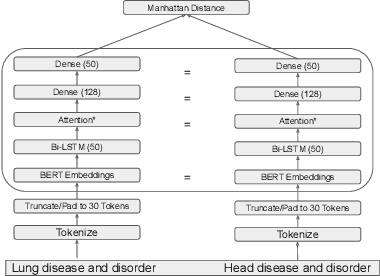
Abstract:The current UMLS (Unified Medical Language System) Metathesaurus construction process for integrating over 200 biomedical source vocabularies is expensive and error-prone as it relies on the lexical algorithms and human editors for deciding if the two biomedical terms are synonymous. Recent advances in Natural Language Processing such as Transformer models like BERT and its biomedical variants with contextualized word embeddings have achieved state-of-the-art (SOTA) performance on downstream tasks. We aim to validate if these approaches using the BERT models can actually outperform the existing approaches for predicting synonymy in the UMLS Metathesaurus. In the existing Siamese Networks with LSTM and BioWordVec embeddings, we replace the BioWordVec embeddings with the biomedical BERT embeddings extracted from each BERT model using different ways of extraction. In the Transformer architecture, we evaluate the use of the different biomedical BERT models that have been pre-trained using different datasets and tasks. Given the SOTA performance of these BERT models for other downstream tasks, our experiments yield surprisingly interesting results: (1) in both model architectures, the approaches employing these biomedical BERT-based models do not outperform the existing approaches using Siamese Network with BioWordVec embeddings for the UMLS synonymy prediction task, (2) the original BioBERT large model that has not been pre-trained with the UMLS outperforms the SapBERT models that have been pre-trained with the UMLS, and (3) using the Siamese Networks yields better performance for synonymy prediction when compared to using the biomedical BERT models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge