Hong Yung Yip
RESTORE: Graph Embedding Assessment Through Reconstruction
Sep 05, 2023Abstract:Following the success of Word2Vec embeddings, graph embeddings (GEs) have gained substantial traction. GEs are commonly generated and evaluated extrinsically on downstream applications, but intrinsic evaluations of the original graph properties in terms of topological structure and semantic information have been lacking. Understanding these will help identify the deficiency of the various families of GE methods when vectorizing graphs in terms of preserving the relevant knowledge or learning incorrect knowledge. To address this, we propose RESTORE, a framework for intrinsic GEs assessment through graph reconstruction. We show that reconstructing the original graph from the underlying GEs yields insights into the relative amount of information preserved in a given vector form. We first introduce the graph reconstruction task. We generate GEs from three GE families based on factorization methods, random walks, and deep learning (with representative algorithms from each family) on the CommonSense Knowledge Graph (CSKG). We analyze their effectiveness in preserving the (a) topological structure of node-level graph reconstruction with an increasing number of hops and (b) semantic information on various word semantic and analogy tests. Our evaluations show deep learning-based GE algorithm (SDNE) is overall better at preserving (a) with a mean average precision (mAP) of 0.54 and 0.35 for 2 and 3-hop reconstruction respectively, while the factorization-based algorithm (HOPE) is better at encapsulating (b) with an average Euclidean distance of 0.14, 0.17, and 0.11 for 1, 2, and 3-hop reconstruction respectively. The modest performance of these GEs leaves room for further research avenues on better graph representation learning.
UBERT: A Novel Language Model for Synonymy Prediction at Scale in the UMLS Metathesaurus
Apr 27, 2022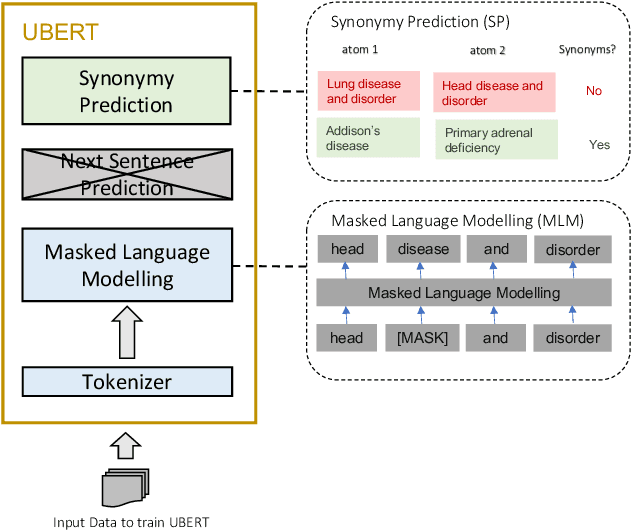

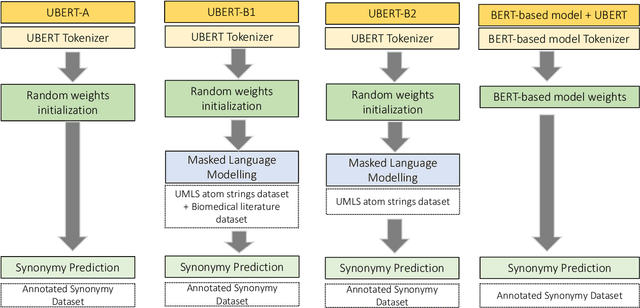

Abstract:The UMLS Metathesaurus integrates more than 200 biomedical source vocabularies. During the Metathesaurus construction process, synonymous terms are clustered into concepts by human editors, assisted by lexical similarity algorithms. This process is error-prone and time-consuming. Recently, a deep learning model (LexLM) has been developed for the UMLS Vocabulary Alignment (UVA) task. This work introduces UBERT, a BERT-based language model, pretrained on UMLS terms via a supervised Synonymy Prediction (SP) task replacing the original Next Sentence Prediction (NSP) task. The effectiveness of UBERT for UMLS Metathesaurus construction process is evaluated using the UMLS Vocabulary Alignment (UVA) task. We show that UBERT outperforms the LexLM, as well as biomedical BERT-based models. Key to the performance of UBERT are the synonymy prediction task specifically developed for UBERT, the tight alignment of training data to the UVA task, and the similarity of the models used for pretrained UBERT.
Evaluating Biomedical BERT Models for Vocabulary Alignment at Scale in the UMLS Metathesaurus
Sep 14, 2021
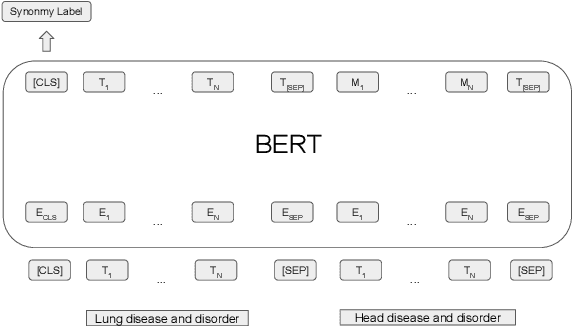

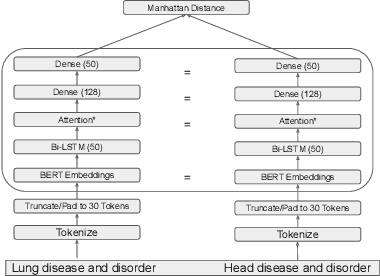
Abstract:The current UMLS (Unified Medical Language System) Metathesaurus construction process for integrating over 200 biomedical source vocabularies is expensive and error-prone as it relies on the lexical algorithms and human editors for deciding if the two biomedical terms are synonymous. Recent advances in Natural Language Processing such as Transformer models like BERT and its biomedical variants with contextualized word embeddings have achieved state-of-the-art (SOTA) performance on downstream tasks. We aim to validate if these approaches using the BERT models can actually outperform the existing approaches for predicting synonymy in the UMLS Metathesaurus. In the existing Siamese Networks with LSTM and BioWordVec embeddings, we replace the BioWordVec embeddings with the biomedical BERT embeddings extracted from each BERT model using different ways of extraction. In the Transformer architecture, we evaluate the use of the different biomedical BERT models that have been pre-trained using different datasets and tasks. Given the SOTA performance of these BERT models for other downstream tasks, our experiments yield surprisingly interesting results: (1) in both model architectures, the approaches employing these biomedical BERT-based models do not outperform the existing approaches using Siamese Network with BioWordVec embeddings for the UMLS synonymy prediction task, (2) the original BioBERT large model that has not been pre-trained with the UMLS outperforms the SapBERT models that have been pre-trained with the UMLS, and (3) using the Siamese Networks yields better performance for synonymy prediction when compared to using the biomedical BERT models.
How will the Internet of Things enable Augmented Personalized Health?
Dec 31, 2017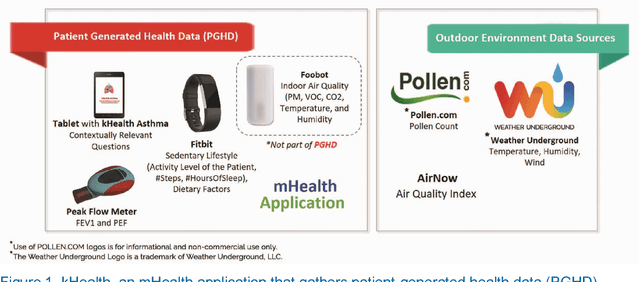
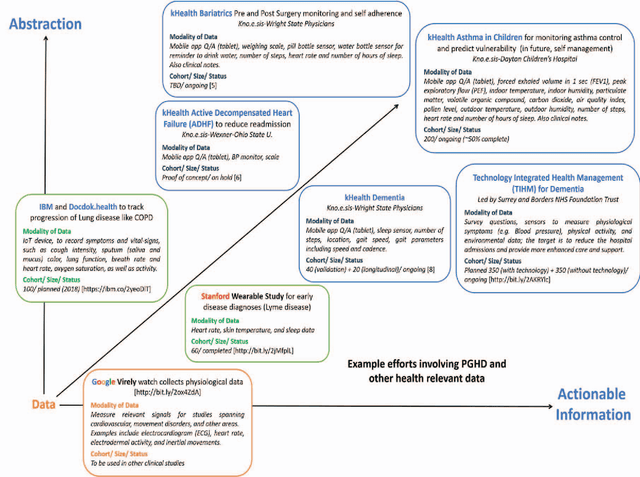
Abstract:Internet-of-Things (IoT) is profoundly redefining the way we create, consume, and share information. Health aficionados and citizens are increasingly using IoT technologies to track their sleep, food intake, activity, vital body signals, and other physiological observations. This is complemented by IoT systems that continuously collect health-related data from the environment and inside the living quarters. Together, these have created an opportunity for a new generation of healthcare solutions. However, interpreting data to understand an individual's health is challenging. It is usually necessary to look at that individual's clinical record and behavioral information, as well as social and environmental information affecting that individual. Interpreting how well a patient is doing also requires looking at his adherence to respective health objectives, application of relevant clinical knowledge and the desired outcomes. We resort to the vision of Augmented Personalized Healthcare (APH) to exploit the extensive variety of relevant data and medical knowledge using Artificial Intelligence (AI) techniques to extend and enhance human health to presents various stages of augmented health management strategies: self-monitoring, self-appraisal, self-management, intervention, and disease progress tracking and prediction. kHealth technology, a specific incarnation of APH, and its application to Asthma and other diseases are used to provide illustrations and discuss alternatives for technology-assisted health management. Several prominent efforts involving IoT and patient-generated health data (PGHD) with respect converting multimodal data into actionable information (big data to smart data) are also identified. Roles of three components in an evidence-based semantic perception approach- Contextualization, Abstraction, and Personalization are discussed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge