Braja Gopal Patra
Extracting Social Support and Social Isolation Information from Clinical Psychiatry Notes: Comparing a Rule-based NLP System and a Large Language Model
Mar 25, 2024Abstract:Background: Social support (SS) and social isolation (SI) are social determinants of health (SDOH) associated with psychiatric outcomes. In electronic health records (EHRs), individual-level SS/SI is typically documented as narrative clinical notes rather than structured coded data. Natural language processing (NLP) algorithms can automate the otherwise labor-intensive process of data extraction. Data and Methods: Psychiatric encounter notes from Mount Sinai Health System (MSHS, n=300) and Weill Cornell Medicine (WCM, n=225) were annotated and established a gold standard corpus. A rule-based system (RBS) involving lexicons and a large language model (LLM) using FLAN-T5-XL were developed to identify mentions of SS and SI and their subcategories (e.g., social network, instrumental support, and loneliness). Results: For extracting SS/SI, the RBS obtained higher macro-averaged f-scores than the LLM at both MSHS (0.89 vs. 0.65) and WCM (0.85 vs. 0.82). For extracting subcategories, the RBS also outperformed the LLM at both MSHS (0.90 vs. 0.62) and WCM (0.82 vs. 0.81). Discussion and Conclusion: Unexpectedly, the RBS outperformed the LLMs across all metrics. Intensive review demonstrates that this finding is due to the divergent approach taken by the RBS and LLM. The RBS were designed and refined to follow the same specific rules as the gold standard annotations. Conversely, the LLM were more inclusive with categorization and conformed to common English-language understanding. Both approaches offer advantages and are made available open-source for future testing.
SODA: A Natural Language Processing Package to Extract Social Determinants of Health for Cancer Studies
Dec 06, 2022



Abstract:Objective: We aim to develop an open-source natural language processing (NLP) package, SODA (i.e., SOcial DeterminAnts), with pre-trained transformer models to extract social determinants of health (SDoH) for cancer patients, examine the generalizability of SODA to a new disease domain (i.e., opioid use), and evaluate the extraction rate of SDoH using cancer populations. Methods: We identified SDoH categories and attributes and developed an SDoH corpus using clinical notes from a general cancer cohort. We compared four transformer-based NLP models to extract SDoH, examined the generalizability of NLP models to a cohort of patients prescribed with opioids, and explored customization strategies to improve performance. We applied the best NLP model to extract 19 categories of SDoH from the breast (n=7,971), lung (n=11,804), and colorectal cancer (n=6,240) cohorts. Results and Conclusion: We developed a corpus of 629 cancer patients notes with annotations of 13,193 SDoH concepts/attributes from 19 categories of SDoH. The Bidirectional Encoder Representations from Transformers (BERT) model achieved the best strict/lenient F1 scores of 0.9216 and 0.9441 for SDoH concept extraction, 0.9617 and 0.9626 for linking attributes to SDoH concepts. Fine-tuning the NLP models using new annotations from opioid use patients improved the strict/lenient F1 scores from 0.8172/0.8502 to 0.8312/0.8679. The extraction rates among 19 categories of SDoH varied greatly, where 10 SDoH could be extracted from >70% of cancer patients, but 9 SDoH had a low extraction rate (<70% of cancer patients). The SODA package with pre-trained transformer models is publicly available at https://github.com/uf-hobiinformatics-lab/SDoH_SODA.
Classifying Cyber-Risky Clinical Notes by Employing Natural Language Processing
Mar 24, 2022


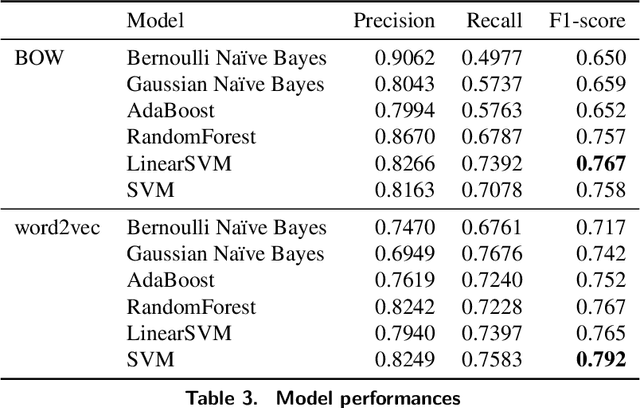
Abstract:Clinical notes, which can be embedded into electronic medical records, document patient care delivery and summarize interactions between healthcare providers and patients. These clinical notes directly inform patient care and can also indirectly inform research and quality/safety metrics, among other indirect metrics. Recently, some states within the United States of America require patients to have open access to their clinical notes to improve the exchange of patient information for patient care. Thus, developing methods to assess the cyber risks of clinical notes before sharing and exchanging data is critical. While existing natural language processing techniques are geared to de-identify clinical notes, to the best of our knowledge, few have focused on classifying sensitive-information risk, which is a fundamental step toward developing effective, widespread protection of patient health information. To bridge this gap, this research investigates methods for identifying security/privacy risks within clinical notes. The classification either can be used upstream to identify areas within notes that likely contain sensitive information or downstream to improve the identification of clinical notes that have not been entirely de-identified. We develop several models using unigram and word2vec features with different classifiers to categorize sentence risk. Experiments on i2b2 de-identification dataset show that the SVM classifier using word2vec features obtained a maximum F1-score of 0.792. Future research involves articulation and differentiation of risk in terms of different global regulatory requirements.
Sentiment Analysis of Code-Mixed Indian Languages: An Overview of SAIL_Code-Mixed Shared Task @ICON-2017
Mar 18, 2018

Abstract:Sentiment analysis is essential in many real-world applications such as stance detection, review analysis, recommendation system, and so on. Sentiment analysis becomes more difficult when the data is noisy and collected from social media. India is a multilingual country; people use more than one languages to communicate within themselves. The switching in between the languages is called code-switching or code-mixing, depending upon the type of mixing. This paper presents overview of the shared task on sentiment analysis of code-mixed data pairs of Hindi-English and Bengali-English collected from the different social media platform. The paper describes the task, dataset, evaluation, baseline and participant's systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge