Developing a general-purpose clinical language inference model from a large corpus of clinical notes
Paper and Code
Oct 12, 2022
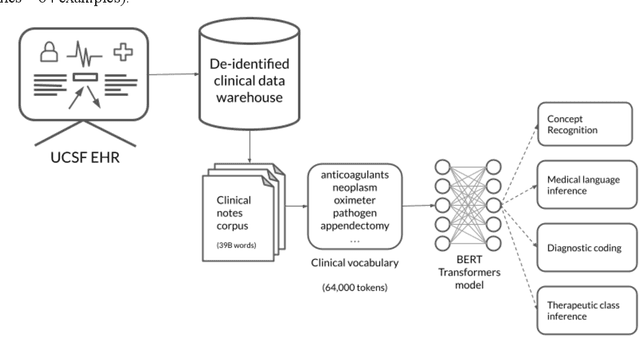
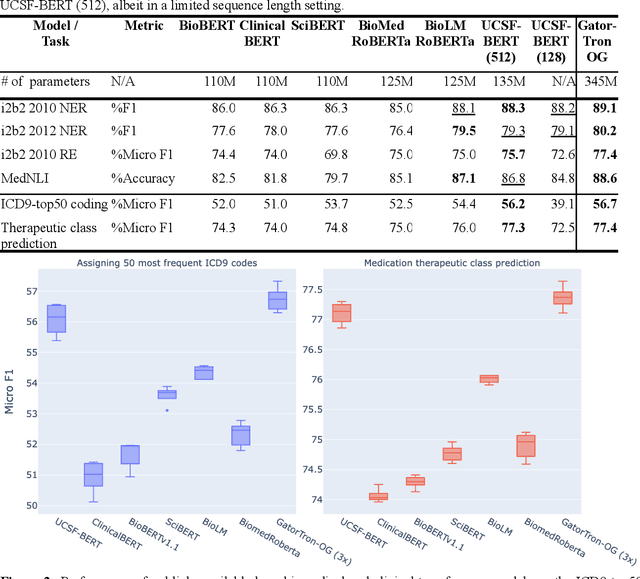
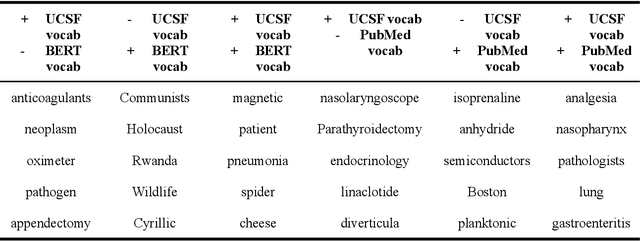
Several biomedical language models have already been developed for clinical language inference. However, these models typically utilize general vocabularies and are trained on relatively small clinical corpora. We sought to evaluate the impact of using a domain-specific vocabulary and a large clinical training corpus on the performance of these language models in clinical language inference. We trained a Bidirectional Encoder Decoder from Transformers (BERT) model using a diverse, deidentified corpus of 75 million deidentified clinical notes authored at the University of California, San Francisco (UCSF). We evaluated this model on several clinical language inference benchmark tasks: clinical and temporal concept recognition, relation extraction and medical language inference. We also evaluated our model on two tasks using discharge summaries from UCSF: diagnostic code assignment and therapeutic class inference. Our model performs at par with the best publicly available biomedical language models of comparable sizes on the public benchmark tasks, and is significantly better than these models in a within-system evaluation on the two tasks using UCSF data. The use of in-domain vocabulary appears to improve the encoding of longer documents. The use of large clinical corpora appears to enhance document encoding and inferential accuracy. However, further research is needed to improve abbreviation resolution, and numerical, temporal, and implicitly causal inference.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge