Jean-Pierre R. Falet
Debiasing Counterfactuals In the Presence of Spurious Correlations
Aug 21, 2023



Abstract:Deep learning models can perform well in complex medical imaging classification tasks, even when basing their conclusions on spurious correlations (i.e. confounders), should they be prevalent in the training dataset, rather than on the causal image markers of interest. This would thereby limit their ability to generalize across the population. Explainability based on counterfactual image generation can be used to expose the confounders but does not provide a strategy to mitigate the bias. In this work, we introduce the first end-to-end training framework that integrates both (i) popular debiasing classifiers (e.g. distributionally robust optimization (DRO)) to avoid latching onto the spurious correlations and (ii) counterfactual image generation to unveil generalizable imaging markers of relevance to the task. Additionally, we propose a novel metric, Spurious Correlation Latching Score (SCLS), to quantify the extent of the classifier reliance on the spurious correlation as exposed by the counterfactual images. Through comprehensive experiments on two public datasets (with the simulated and real visual artifacts), we demonstrate that the debiasing method: (i) learns generalizable markers across the population, and (ii) successfully ignores spurious correlations and focuses on the underlying disease pathology.
Clinically Plausible Pathology-Anatomy Disentanglement in Patient Brain MRI with Structured Variational Priors
Nov 16, 2022


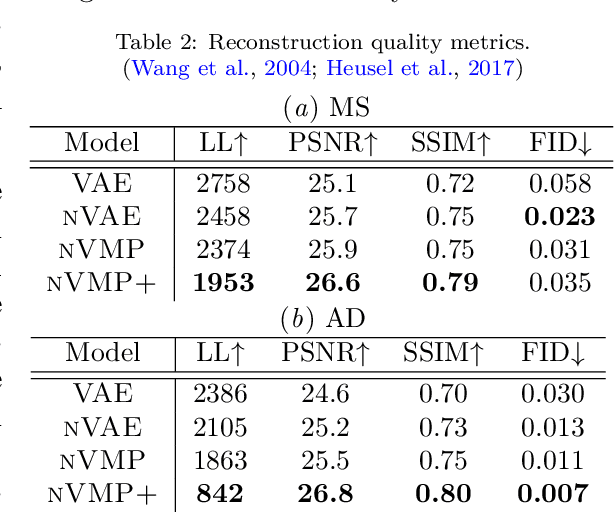
Abstract:We propose a hierarchically structured variational inference model for accurately disentangling observable evidence of disease (e.g. brain lesions or atrophy) from subject-specific anatomy in brain MRIs. With flexible, partially autoregressive priors, our model (1) addresses the subtle and fine-grained dependencies that typically exist between anatomical and pathological generating factors of an MRI to ensure the clinical validity of generated samples; (2) preserves and disentangles finer pathological details pertaining to a patient's disease state. Additionally, we experiment with an alternative training configuration where we provide supervision to a subset of latent units. It is shown that (1) a partially supervised latent space achieves a higher degree of disentanglement between evidence of disease and subject-specific anatomy; (2) when the prior is formulated with an autoregressive structure, knowledge from the supervision can propagate to the unsupervised latent units, resulting in more informative latent representations capable of modelling anatomy-pathology interdependencies.
Rethinking Generalization: The Impact of Annotation Style on Medical Image Segmentation
Oct 31, 2022Abstract:Generalization is an important attribute of machine learning models, particularly for those that are to be deployed in a medical context, where unreliable predictions can have real world consequences. While the failure of models to generalize across datasets is typically attributed to a mismatch in the data distributions, performance gaps are often a consequence of biases in the ``ground-truth" label annotations. This is particularly important in the context of medical image segmentation of pathological structures (e.g. lesions), where the annotation process is much more subjective, and affected by a number underlying factors, including the annotation protocol, rater education/experience, and clinical aims, among others. In this paper, we show that modeling annotation biases, rather than ignoring them, poses a promising way of accounting for differences in annotation style across datasets. To this end, we propose a generalized conditioning framework to (1) learn and account for different annotation styles across multiple datasets using a single model, (2) identify similar annotation styles across different datasets in order to permit their effective aggregation, and (3) fine-tune a fully trained model to a new annotation style with just a few samples. Next, we present an image-conditioning approach to model annotation styles that correlate with specific image features, potentially enabling detection biases to be more easily identified.
Counterfactual Image Synthesis for Discovery of Personalized Predictive Image Markers
Aug 03, 2022



Abstract:The discovery of patient-specific imaging markers that are predictive of future disease outcomes can help us better understand individual-level heterogeneity of disease evolution. In fact, deep learning models that can provide data-driven personalized markers are much more likely to be adopted in medical practice. In this work, we demonstrate that data-driven biomarker discovery can be achieved through a counterfactual synthesis process. We show how a deep conditional generative model can be used to perturb local imaging features in baseline images that are pertinent to subject-specific future disease evolution and result in a counterfactual image that is expected to have a different future outcome. Candidate biomarkers, therefore, result from examining the set of features that are perturbed in this process. Through several experiments on a large-scale, multi-scanner, multi-center multiple sclerosis (MS) clinical trial magnetic resonance imaging (MRI) dataset of relapsing-remitting (RRMS) patients, we demonstrate that our model produces counterfactuals with changes in imaging features that reflect established clinical markers predictive of future MRI lesional activity at the population level. Additional qualitative results illustrate that our model has the potential to discover novel and subject-specific predictive markers of future activity.
Personalized Prediction of Future Lesion Activity and Treatment Effect in Multiple Sclerosis from Baseline MRI
Apr 01, 2022



Abstract:Precision medicine for chronic diseases such as multiple sclerosis (MS) involves choosing a treatment which best balances efficacy and side effects/preferences for individual patients. Making this choice as early as possible is important, as delays in finding an effective therapy can lead to irreversible disability accrual. To this end, we present the first deep neural network model for individualized treatment decisions from baseline magnetic resonance imaging (MRI) (with clinical information if available) for MS patients. Our model (a) predicts future new and enlarging T2 weighted (NE-T2) lesion counts on follow-up MRI on multiple treatments and (b) estimates the conditional average treatment effect (CATE), as defined by the predicted future suppression of NE-T2 lesions, between different treatment options relative to placebo. Our model is validated on a proprietary federated dataset of 1817 multi-sequence MRIs acquired from MS patients during four multi-centre randomized clinical trials. Our framework achieves high average precision in the binarized regression of future NE-T2 lesions on five different treatments, identifies heterogeneous treatment effects, and provides a personalized treatment recommendation that accounts for treatment-associated risk (e.g. side effects, patient preference, administration difficulties).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge