Leonid Mill
DiffRenderGAN: Addressing Training Data Scarcity in Deep Segmentation Networks for Quantitative Nanomaterial Analysis through Differentiable Rendering and Generative Modelling
Feb 13, 2025Abstract:Nanomaterials exhibit distinctive properties governed by parameters such as size, shape, and surface characteristics, which critically influence their applications and interactions across technological, biological, and environmental contexts. Accurate quantification and understanding of these materials are essential for advancing research and innovation. In this regard, deep learning segmentation networks have emerged as powerful tools that enable automated insights and replace subjective methods with precise quantitative analysis. However, their efficacy depends on representative annotated datasets, which are challenging to obtain due to the costly imaging of nanoparticles and the labor-intensive nature of manual annotations. To overcome these limitations, we introduce DiffRenderGAN, a novel generative model designed to produce annotated synthetic data. By integrating a differentiable renderer into a Generative Adversarial Network (GAN) framework, DiffRenderGAN optimizes textural rendering parameters to generate realistic, annotated nanoparticle images from non-annotated real microscopy images. This approach reduces the need for manual intervention and enhances segmentation performance compared to existing synthetic data methods by generating diverse and realistic data. Tested on multiple ion and electron microscopy cases, including titanium dioxide (TiO$_2$), silicon dioxide (SiO$_2$)), and silver nanowires (AgNW), DiffRenderGAN bridges the gap between synthetic and real data, advancing the quantification and understanding of complex nanomaterial systems.
SYNTA: A novel approach for deep learning-based image analysis in muscle histopathology using photo-realistic synthetic data
Aug 03, 2022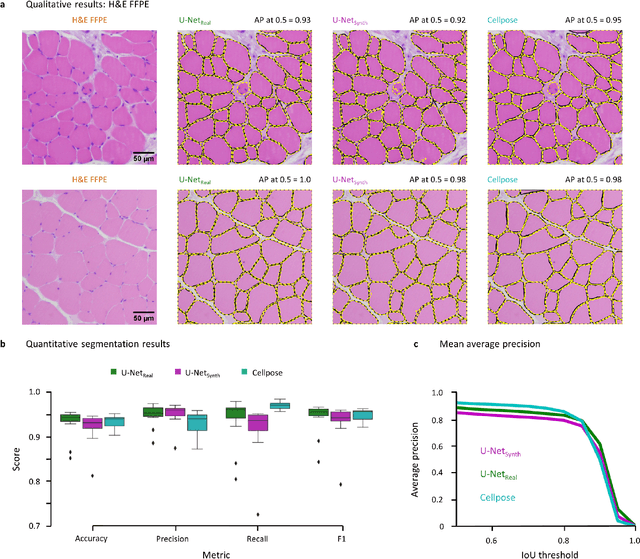
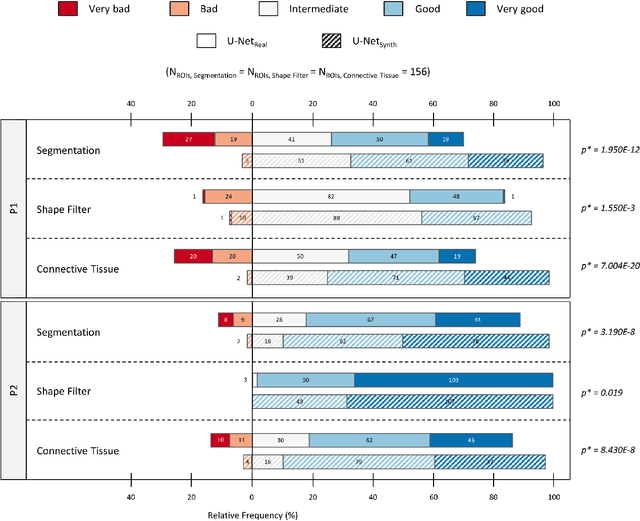
Abstract:Artificial intelligence (AI), machine learning, and deep learning (DL) methods are becoming increasingly important in the field of biomedical image analysis. However, to exploit the full potential of such methods, a representative number of experimentally acquired images containing a significant number of manually annotated objects is needed as training data. Here we introduce SYNTA (synthetic data) as a novel approach for the generation of synthetic, photo-realistic, and highly complex biomedical images as training data for DL systems. We show the versatility of our approach in the context of muscle fiber and connective tissue analysis in histological sections. We demonstrate that it is possible to perform robust and expert-level segmentation tasks on previously unseen real-world data, without the need for manual annotations using synthetic training data alone. Being a fully parametric technique, our approach poses an interpretable and controllable alternative to Generative Adversarial Networks (GANs) and has the potential to significantly accelerate quantitative image analysis in a variety of biomedical applications in microscopy and beyond.
Synthetic Image Rendering Solves Annotation Problem in Deep Learning Nanoparticle Segmentation
Nov 20, 2020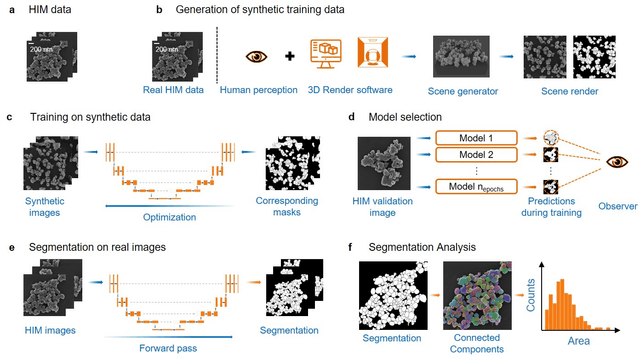
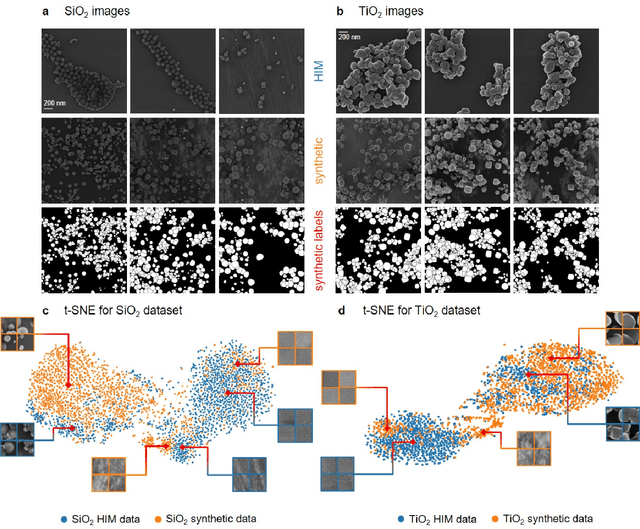
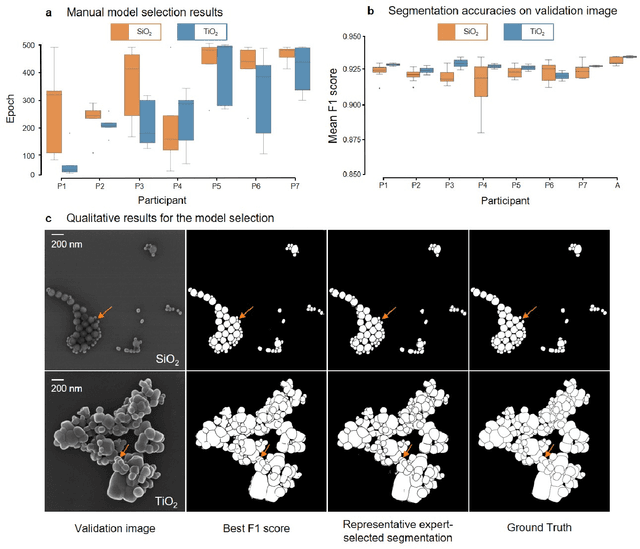
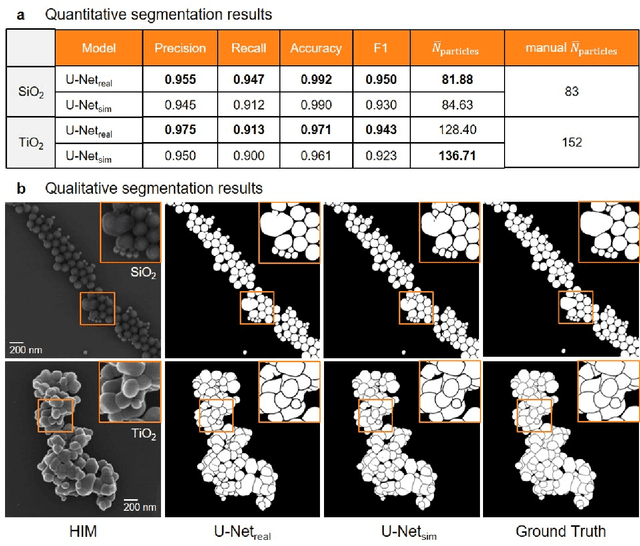
Abstract:Nanoparticles occur in various environments as a consequence of man-made processes, which raises concerns about their impact on the environment and human health. To allow for proper risk assessment, a precise and statistically relevant analysis of particle characteristics (such as e.g. size, shape and composition) is required that would greatly benefit from automated image analysis procedures. While deep learning shows impressive results in object detection tasks, its applicability is limited by the amount of representative, experimentally collected and manually annotated training data. Here, we present an elegant, flexible and versatile method to bypass this costly and tedious data acquisition process. We show that using a rendering software allows to generate realistic, synthetic training data to train a state-of-the art deep neural network. Using this approach, we derive a segmentation accuracy that is comparable to man-made annotations for toxicologically relevant metal-oxide nanoparticle ensembles which we chose as examples. Our study paves the way towards the use of deep learning for automated, high-throughput particle detection in a variety of imaging techniques such as microscopies and spectroscopies, for a wide variety of studies and applications, including the detection of plastic micro- and nanoparticles.
Cephalogram Synthesis and Landmark Detection in Dental Cone-Beam CT Systems
Sep 09, 2020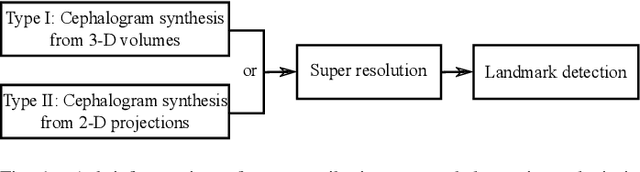
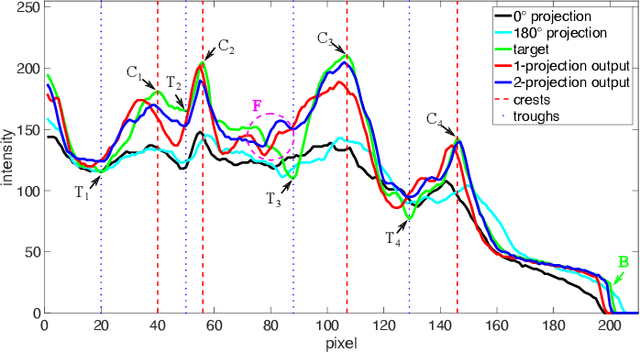
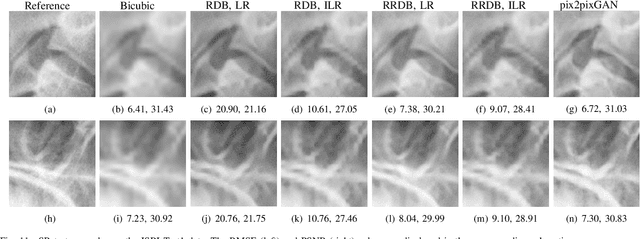
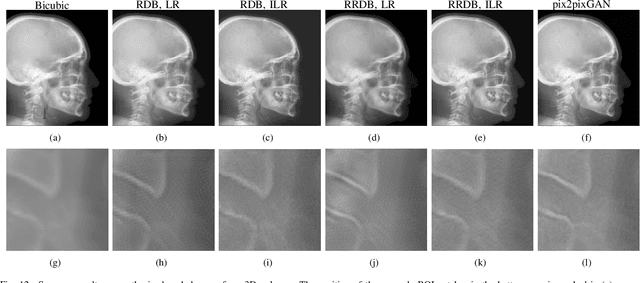
Abstract:Due to the lack of standardized 3D cephalometric analytic methodology, 2D cephalograms synthesized from 3D cone-beam computed tomography (CBCT) volumes are widely used for cephalometric analysis in dental CBCT systems. However, compared with conventional X-ray film based cephalograms, such synthetic cephalograms lack image contrast and resolution. In addition, the radiation dose during the scan for 3D reconstruction causes potential health risks. In this work, we propose a sigmoid-based intensity transform that uses the nonlinear optical property of X-ray films to increase image contrast of synthetic cephalograms. To improve image resolution, super resolution deep learning techniques are investigated. For low dose purpose, the pixel-to-pixel generative adversarial network (pix2pixGAN) is proposed for 2D cephalogram synthesis directly from two CBCT projections. For landmark detection in the synthetic cephalograms, an efficient automatic landmark detection method using the combination of LeNet-5 and ResNet50 is proposed. Our experiments demonstrate the efficacy of pix2pixGAN in 2D cephalogram synthesis, achieving an average peak signal-to-noise ratio (PSNR) value of 33.8 with reference to the cephalograms synthesized from 3D CBCT volumes. Pix2pixGAN also achieves the best performance in super resolution, achieving an average PSNR value of 32.5 without the introduction of checkerboard or jagging artifacts. Our proposed automatic landmark detection method achieves 86.7% successful detection rate in the 2 mm clinical acceptable range on the ISBI Test1 data, which is comparable to the state-of-the-art methods. The method trained on conventional cephalograms can be directly applied to landmark detection in the synthetic cephalograms, achieving 93.0% and 80.7% successful detection rate in 4 mm precision range for synthetic cephalograms from 3D volumes and 2D projections respectively.
Learning with Known Operators reduces Maximum Training Error Bounds
Jul 03, 2019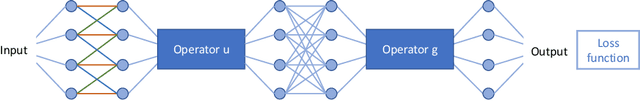
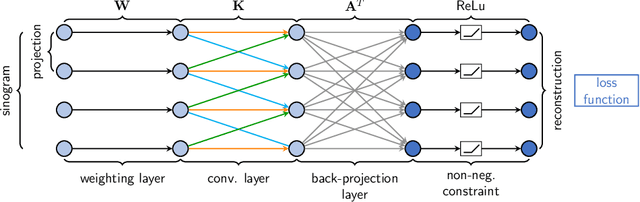
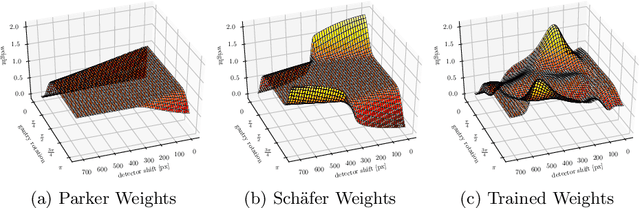
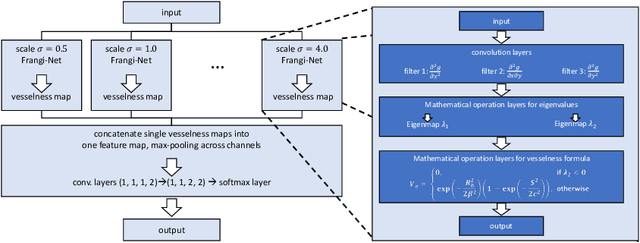
Abstract:We describe an approach for incorporating prior knowledge into machine learning algorithms. We aim at applications in physics and signal processing in which we know that certain operations must be embedded into the algorithm. Any operation that allows computation of a gradient or sub-gradient towards its inputs is suited for our framework. We derive a maximal error bound for deep nets that demonstrates that inclusion of prior knowledge results in its reduction. Furthermore, we also show experimentally that known operators reduce the number of free parameters. We apply this approach to various tasks ranging from CT image reconstruction over vessel segmentation to the derivation of previously unknown imaging algorithms. As such the concept is widely applicable for many researchers in physics, imaging, and signal processing. We assume that our analysis will support further investigation of known operators in other fields of physics, imaging, and signal processing.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge