Andre Aichert
on behalf of EUCanImage working group
Robust Multicentre Detection and Classification of Colorectal Liver Metastases on CT: Application of Foundation Models
Jan 12, 2026Abstract:Colorectal liver metastases (CRLM) are a major cause of cancer-related mortality, and reliable detection on CT remains challenging in multi-centre settings. We developed a foundation model-based AI pipeline for patient-level classification and lesion-level detection of CRLM on contrast-enhanced CT, integrating uncertainty quantification and explainability. CT data from the EuCanImage consortium (n=2437) and an external TCIA cohort (n=197) were used. Among several pretrained models, UMedPT achieved the best performance and was fine-tuned with an MLP head for classification and an FCOS-based head for lesion detection. The classification model achieved an AUC of 0.90 and a sensitivity of 0.82 on the combined test set, with a sensitivity of 0.85 on the external cohort. Excluding the most uncertain 20 percent of cases improved AUC to 0.91 and balanced accuracy to 0.86. Decision curve analysis showed clinical benefit for threshold probabilities between 0.30 and 0.40. The detection model identified 69.1 percent of lesions overall, increasing from 30 percent to 98 percent across lesion size quartiles. Grad-CAM highlighted lesion-corresponding regions in high-confidence cases. These results demonstrate that foundation model-based pipelines can support robust and interpretable CRLM detection and classification across heterogeneous CT data.
End-to-end Learning for Image-based Detection of Molecular Alterations in Digital Pathology
Jun 30, 2022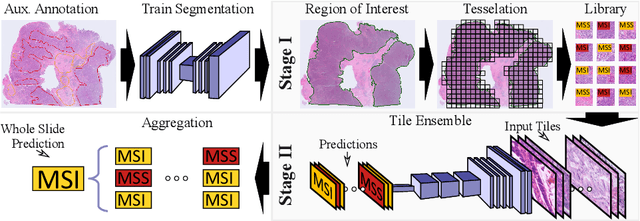
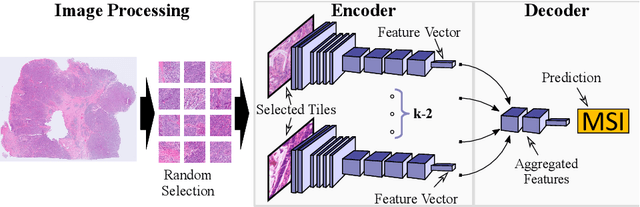
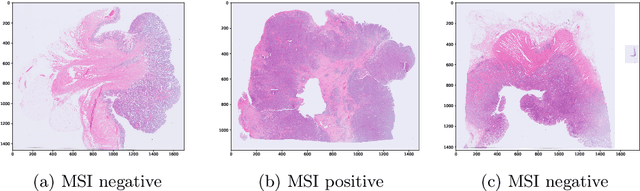
Abstract:Current approaches for classification of whole slide images (WSI) in digital pathology predominantly utilize a two-stage learning pipeline. The first stage identifies areas of interest (e.g. tumor tissue), while the second stage processes cropped tiles from these areas in a supervised fashion. During inference, a large number of tiles are combined into a unified prediction for the entire slide. A major drawback of such approaches is the requirement for task-specific auxiliary labels which are not acquired in clinical routine. We propose a novel learning pipeline for WSI classification that is trainable end-to-end and does not require any auxiliary annotations. We apply our approach to predict molecular alterations for a number of different use-cases, including detection of microsatellite instability in colorectal tumors and prediction of specific mutations for colon, lung, and breast cancer cases from The Cancer Genome Atlas. Results reach AUC scores of up to 94% and are shown to be competitive with state of the art two-stage pipelines. We believe our approach can facilitate future research in digital pathology and contribute to solve a large range of problems around the prediction of cancer phenotypes, hopefully enabling personalized therapies for more patients in future.
Disassemblable Fieldwork CT Scanner Using a 3D-printed Calibration Phantom
Nov 12, 2020



Abstract:The use of computed tomography (CT) imaging has become of increasing interest to academic areas outside of the field of medical imaging and industrial inspection, e.g., to biology and cultural heritage research. The pecularities of these fields, however, sometimes require that objects need to be imaged on-site, e.g., in field-work conditions or in museum collections. Under these circumstances, it is often not possible to use a commercial device and a custom solution is the only viable option. In order to achieve high image quality under adverse conditions, reliable calibration and trajectory reproduction are usually key requirements for any custom CT scanning system. Here, we introduce the construction of a low-cost disassemblable CT scanner that allows calibration even when trajectory reproduction is not possible due to the limitations imposed by the project conditions. Using 3D-printed in-image calibration phantoms, we compute a projection matrix directly from each captured X-ray projection. We describe our method in detail and show successful tomographic reconstructions of several specimen as proof of concept.
* This paper was originally published at the 6th International Conference on Image Formation in X-Ray Computed Tomography (CTmeeting 2020)
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge