Hanibal Bohnenberger
MGPATH: Vision-Language Model with Multi-Granular Prompt Learning for Few-Shot WSI Classification
Feb 11, 2025



Abstract:Whole slide pathology image classification presents challenges due to gigapixel image sizes and limited annotation labels, hindering model generalization. This paper introduces a prompt learning method to adapt large vision-language models for few-shot pathology classification. We first extend the Prov-GigaPath vision foundation model, pre-trained on 1.3 billion pathology image tiles, into a vision-language model by adding adaptors and aligning it with medical text encoders via contrastive learning on 923K image-text pairs. The model is then used to extract visual features and text embeddings from few-shot annotations and fine-tunes with learnable prompt embeddings. Unlike prior methods that combine prompts with frozen features using prefix embeddings or self-attention, we propose multi-granular attention that compares interactions between learnable prompts with individual image patches and groups of them. This approach improves the model's ability to capture both fine-grained details and broader context, enhancing its recognition of complex patterns across sub-regions. To further improve accuracy, we leverage (unbalanced) optimal transport-based visual-text distance to secure model robustness by mitigating perturbations that might occur during the data augmentation process. Empirical experiments on lung, kidney, and breast pathology modalities validate the effectiveness of our approach; thereby, we surpass several of the latest competitors and consistently improve performance across diverse architectures, including CLIP, PLIP, and Prov-GigaPath integrated PLIP. We release our implementations and pre-trained models at this MGPATH.
End-to-end Learning for Image-based Detection of Molecular Alterations in Digital Pathology
Jun 30, 2022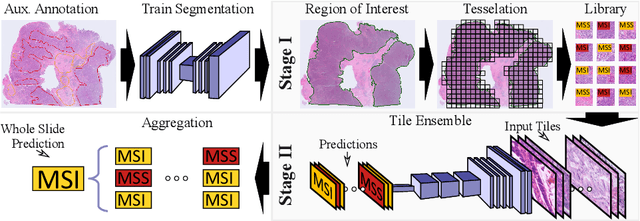
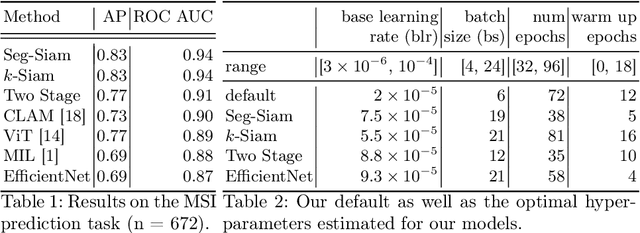
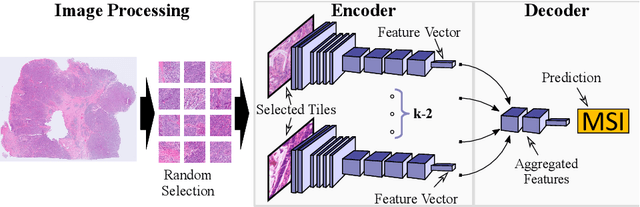
Abstract:Current approaches for classification of whole slide images (WSI) in digital pathology predominantly utilize a two-stage learning pipeline. The first stage identifies areas of interest (e.g. tumor tissue), while the second stage processes cropped tiles from these areas in a supervised fashion. During inference, a large number of tiles are combined into a unified prediction for the entire slide. A major drawback of such approaches is the requirement for task-specific auxiliary labels which are not acquired in clinical routine. We propose a novel learning pipeline for WSI classification that is trainable end-to-end and does not require any auxiliary annotations. We apply our approach to predict molecular alterations for a number of different use-cases, including detection of microsatellite instability in colorectal tumors and prediction of specific mutations for colon, lung, and breast cancer cases from The Cancer Genome Atlas. Results reach AUC scores of up to 94% and are shown to be competitive with state of the art two-stage pipelines. We believe our approach can facilitate future research in digital pathology and contribute to solve a large range of problems around the prediction of cancer phenotypes, hopefully enabling personalized therapies for more patients in future.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge